To apply an alignment highlighting mode, select it in the Highlighting context menu:
| HTML |
|---|
<center>
<br>
<img src="/wiki/download/attachments/54362601/Highlighting Alignment.png"/>
<br>
</center> |
or on the Highlighting tab of the Options Panel:
| HTML |
|---|
<center>
<br>
<img src="/wiki/download/attachments/16122233/Highlighting Alignment_1.png"/>
<br>
</center> |
The following modes are available:
Agreements — highlights symbols that coincide with the reference sequence.
Disagreements — highlights nucleotides that differ from the reference sequence.
- Gaps - highlights gaps.
- Conservation level - highlights conservation level of symbols in a multiple alignment >= or <= treshhold. To select the conservation parameters use the Highlighting Options Panel tab.
- Transitions - highlights transitions.
- Transversions - highlights transversions.
To use dots instead of symbols which are not highlighted check the Use dots checkbox in the Options Panel or use the Highlighting->Use dots context menu item.
To select a reference sequence use the Set this sequence as reference context menu or Reference sequence field in the Highlighting tab of the Options Panel.
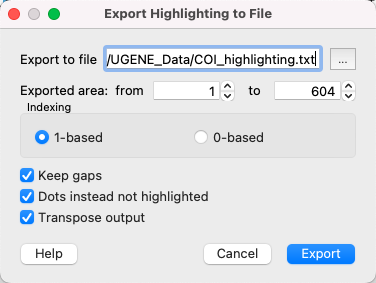
Also you can export highlighting with a help of the Export button in the Options Panel or by the Export->Export highlighted context menu item. The following dialog will appear:
| HTML |
|---|
<center>
<br>
<img src="/wiki/download/attachments/7667818/Highlighting Alignment_2.png"/>
<br>
</center> |
Select file to export, exported area and click on the Export button. The task report will appear in the Notifications
Select the output file to export the highlighting, choose exported area and click on the Export button. The task report will appear in the Notifications.
Also, you may set additional parameters:
- Indexing - select the number to start counting from:
- 1-based - the first column has number 1,
- 0-based - the first column has number 0.
- Keep gaps - check if you want to store gap symbols in the result file.
- Dots instead not highlighted - replace not-highlighted bases with dots in the result file.
- Transpose output - if this checkbox is checked, the sequences will be located horizontally (not vertically, as in UGENE). May be useful, if you have a long alignment, but not many sequences.