Selection of a sequence region is synchronized between different Sequence View components.
Selection in the Sequence View components
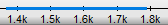
In the Sequence Overview the selection is displayed as a blue line:
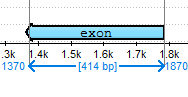
In the Zoom View a blue line with coordinates and the length of the selected region is shown:
In the Details View a rectangle with a dashed border is shown around the selected region:
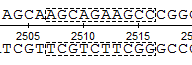
Finally, when one selects an annotation in the Annotations Editor, the corresponding region is also selected in the other components and visa versa.
Selecting a region using the mouse
It is possible to select a region of a sequence using the mouse in each component of the Sequence View.
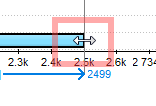
In the Zoom View and the Details View the selection can be adjusted by dragging its left or right border:
Selecting a region using the dialog
If information about exact coordinates of a region is available, click the Select sequence region button on the toolbar or Select–>Sequence region in the context menu of the Sequence View. Input the coordinates in the Region Selection dialog:
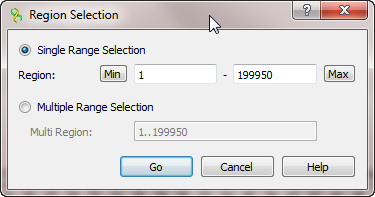
It is possible to input:
- Single Range Selection — a common region with a single range. Use the Min and Max buttons to automatically specify the beginning and the end of the sequence in the corresponding fields.
- Multiple Range Selection — a region that consists of several join elements, for example, exons of a gene. See "The DDBJ/ENA/GenBank Feature Table Definition" for details.
Selecting a region around or between annotations
To select a region between two annotations, for example, an intron region between two exons:
- Click on the annotations, holding the Ctrl/Cmd key:

- Click Select–>Sequence between selected annotations in the context menu of the Sequence View.
To select a region that contains the annotations in a single range, choose the Sequence around selected annotations item in the menu.
