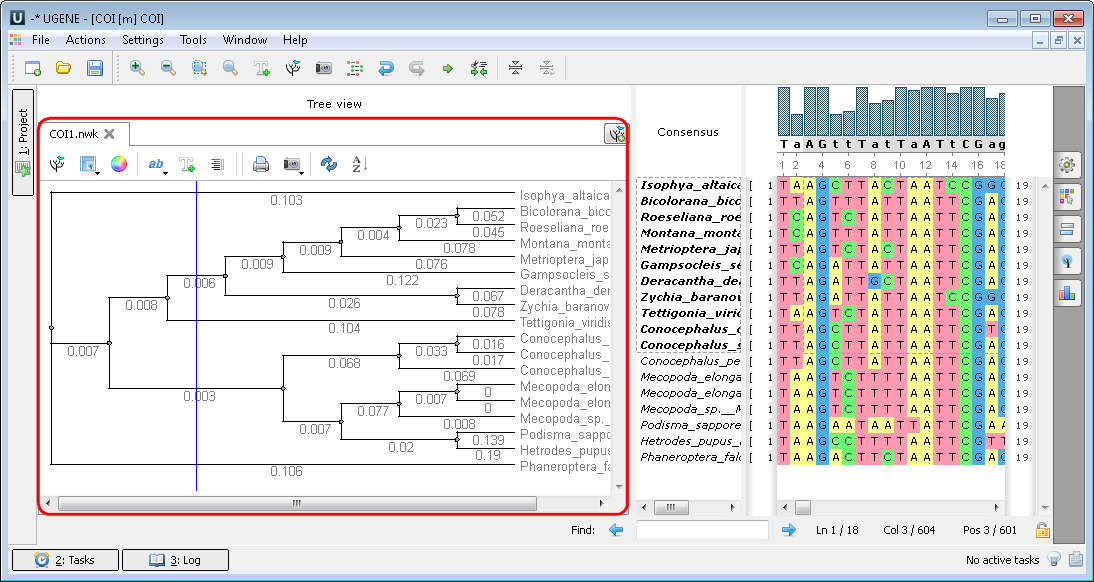
Multiple sequence alignment (MSA Editor). If you open a multiple sequence alignment, for example ugene/data/samples/CLUSTALW/COI.aln, Alignment Editor appears. The following views are available: the project view with your documents, the Alignment Editor with your alignment, options panel with additional info and algorithms:

Alignment Editor - an object view used to visualize and edit DNA, RNA or protein multiple sequence alignments.
You can also use algorithms and instruments from the context menu or the toolbar:
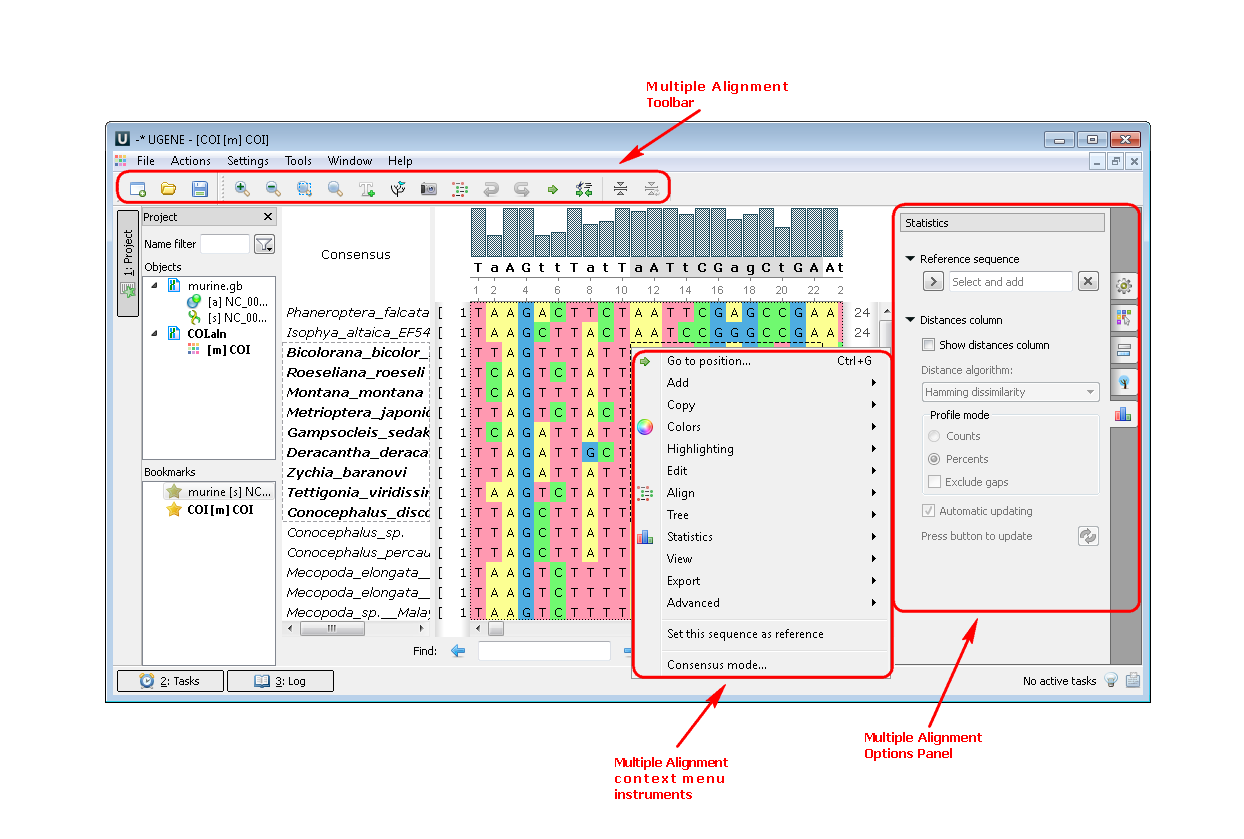
Example 2:Build a tree from your alignment. You can do this by three different ways:
a. From the toolbar:

b. From the context menu:
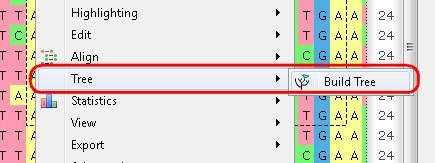
c. From the Options Panel:
After calculation the tree appears in the MSA Editor in a separate window: