The Find Group of Annotated Regions feature provides an algorithm to search for sequence regions that contain a predefined set of annotations.
Open a DNA sequence in the Sequence View. There are two ways to open the Find Repeats dialog:
This tool has been designed to search for annotations that intersect (or completely overlap - it depends on the specified parameters) other, already existing annotations of a given sequence. Let's look at the example: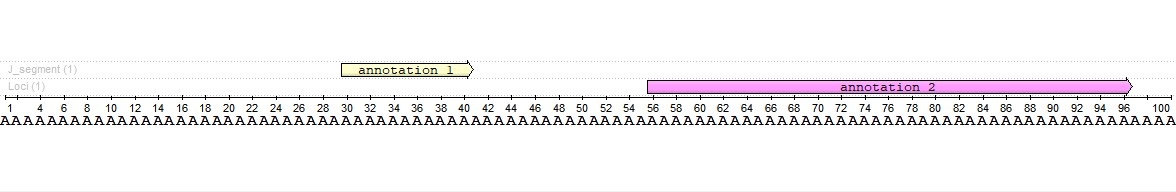
We have a sequence with two annotations. Annotations have different lengths and do not intersect each other. The annotation 2 length is four times the annotation 1 length (41 vs. 11 bases).
Using this function, we can find an annotation that intersects both source annotations and captures their shares depending on their lengths. For example, lets find an intersection 25 bases long. We will have the following annotation:
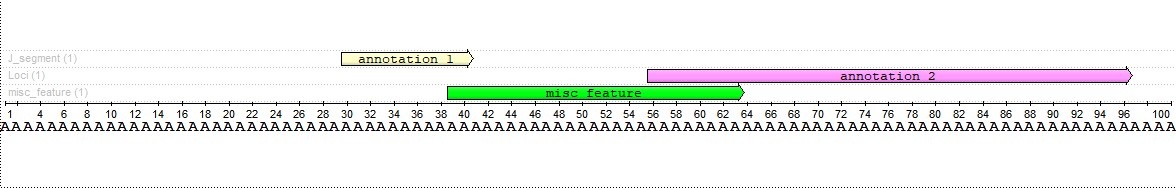
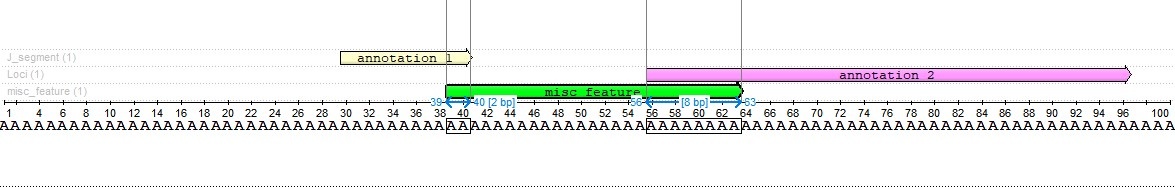
As we can see, the intersection with the first annotation is two characters long, and the intersection with the second annotation is eight characters long. This result was chosen because the second annotation is four times the length of the first annotation.
NOTE: A good candidate for this feature could be any file in Genbank format with a rich set of annotations. FASTA is not the best option, because this format does not store annotations.
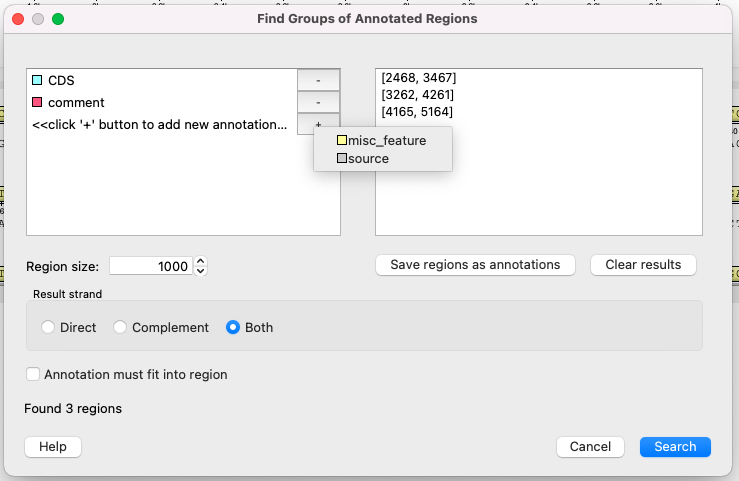 The following parameters are avaliable:
The following parameters are avaliable: