It is possible to apply different coloring schemes for an alignment, so that nucleotide or amino acid residues are colored depending on some rules.
To set another coloring scheme, select the required item in the main (Actions ‣ Colors) or context menu (Colors) of the Alignment Editor, or in the Color combo box on the Highlighting tab on the Options Panel.
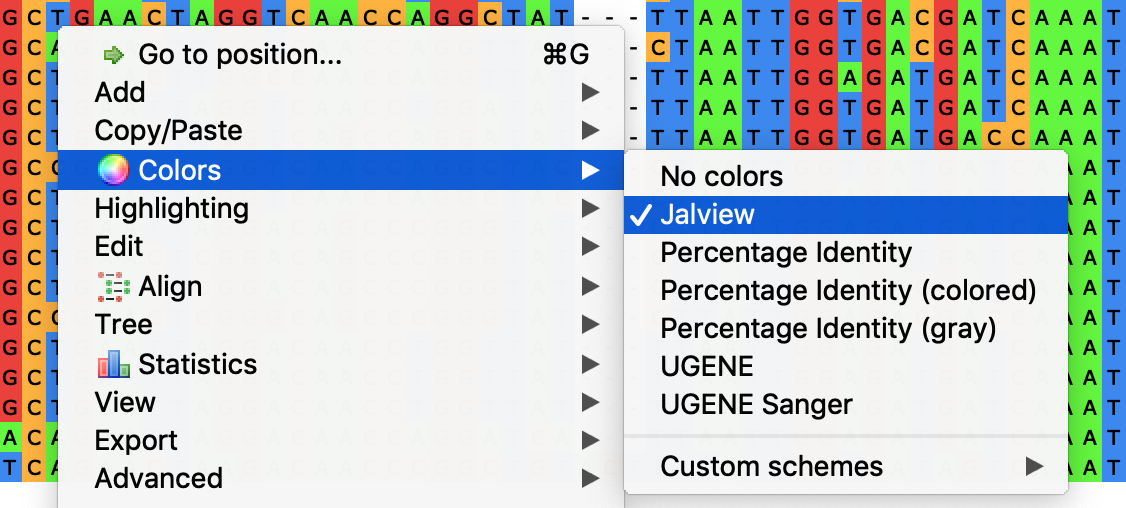
Note that the coloring schemes setting may interfere with other alignment highlighting settings (see e.g. Highlighting combo box on the Highlighting tab of the Options Panel). Thus, the alignment characters colors may vary in this case from the described below.
Coloring Schemes for Nucleotides
The following schemes options are available for a nucleotide alignment:
- No colors — do not color the characters background (i. e. white background is used for all characters).
- Jalview — use the following background colors: A - green, C - orange, G - red, T/U - blue. Gaps and characters of the extended nucleotide alphabet are not colored.
- Percentage Identity — a character background color depends on the character presence in the column of the alignment: below 40% - white, 41-60% - light slate blue, 61-80% - medium slate blue, 81-100% dark slate blue.
- Percentage Identity (colored) — colors depend also on percentage of characters in a column, the following rules are applied (in the order of priority):
- If all characters (100%) are the same and there are no gaps, the characters in the column have yellow background and red font.
- If there are 50% of one character in the column and 50% of another (gaps are not taken into account), one half of the characters has green background and black font (the half to be colored is determined based on the following characters priorities: T>U>G>C>A>B>D>H>K>M>R>S>V>W>Y>N).
- If there is one or several characters in a column with percentage number more or equal to the threshold parameter (see the Threshold parameter on the Highlighting tab of the Options Panel), then most frequent characters are colored as follows: light blue background color and blue font. Note that gaps are considered as characters when calculating percentage number in the column in this case. Also, if required, the same characters priorities are applied as in the previous rule.
- If the rules above are not applied for a character, then it remains uncolored, i.e. white background color and black font are used.
- Percentage Identity (gray) — this coloring scheme is similar to Percentage Identity scheme, but shades of gray color are used instead of the slate blue color.
- UGENE — this is the default coloring scheme, the following background colors are used: A - yellow, C - light green, G - light blue, T/U - light red, gap - white. Other colors may are used for characters of the extended nucleotide alphabet.
- UGENE Sanger — this coloring scheme is used in the UGENE Sanger Reads Editor, the background colors correspond to the peaks of Sanger chromatogram peaks: A - green, C - blue, G - light gray, T - red. The orange background color is used for a gap, magenta background color is used for characters of the extended nucleotide alphabet.
- A custom scheme can be created and used with colors that depend on a character.
Coloring Schemes for Amino Acids
The following schemes options are available for an amino acids alignment:
- No colors — do not color the characters background (i. e. white background is used for all characters).
- Buried index —
- Clustal X —
- Helix propensity —
- Hydrophobicity —
- Percentage Identity —
- Percentage Identity (gray) —
- Strand propensity —
- Tailor —
- Turn propensity —
- UGENE —
- Zappo — amino acids residues are colored according to their physicochemical properties: alphatic/hydrophobic (characters ILVAM) - rose, aromatic (FWY) - orange, positive (KRH) - dark blue, negative (DE) - red, hydrophilic (STNQ) - light green, conformationally special (PG) - magenta, cysteine (C) - yellow.
- A custom scheme can be created and used with colors that depend on a character.