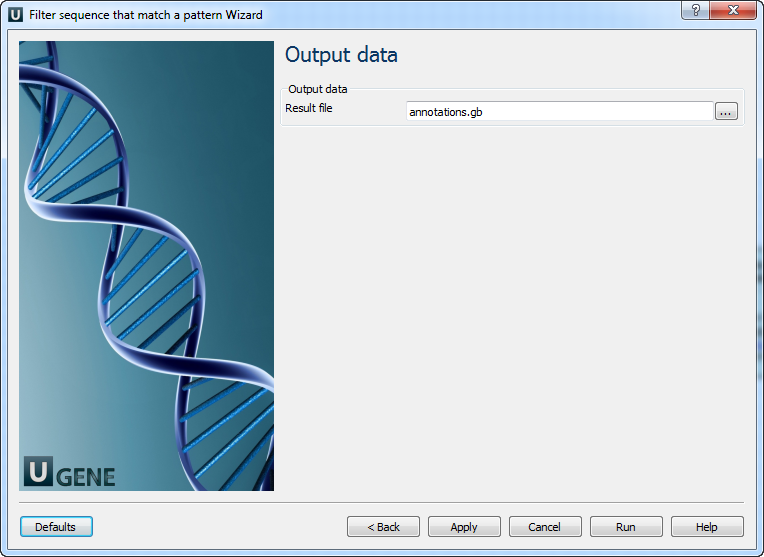
Using thist workflow you can select (or reject) only those sequence that match any pattern you input.
How to Use This Sample
If you haven't used the workflow samples in UGENE before, look at the "How to Use Sample Workflows" section of the documentation.
Workflow Sample Location
The workflow sample "Filter Sequence That Match a Pattern" can be found in the "Scenarios" section of the Workflow Designer samples.
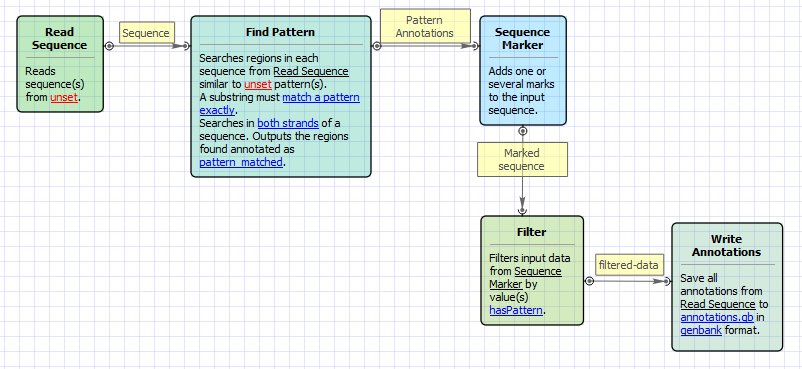
Workflow Image
The workflow looks as follows:
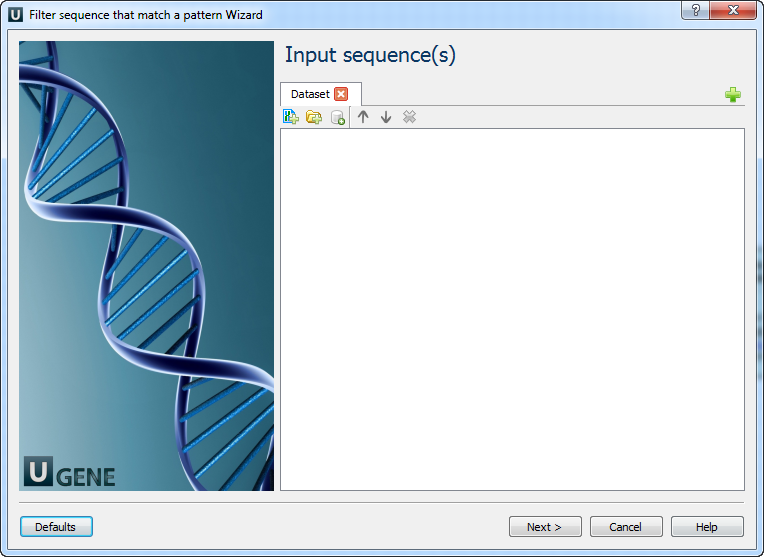
Workflow Wizard
The wizard has 3 pages.
Input sequence(s): On this page you must input sequence(s).
Find pattern: On this page you must input pattern(s) and you can modify searching parameters.
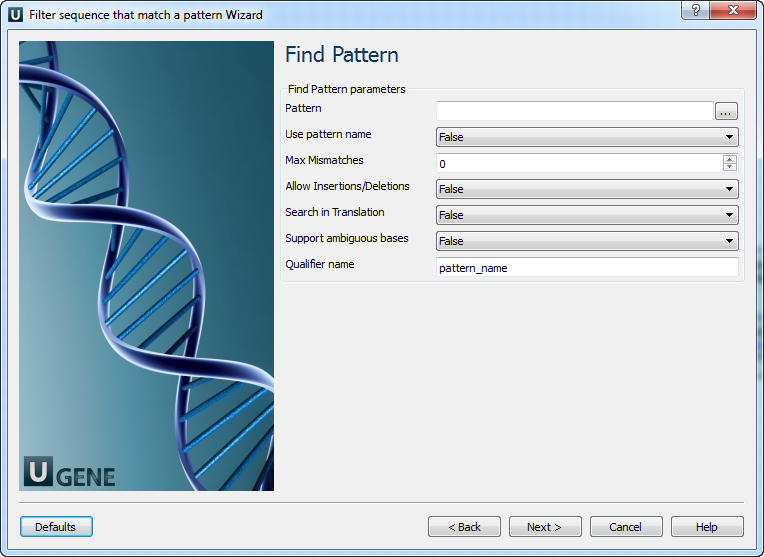
The following parameters are available:
Pattern Semicolon-separated list of patterns to search for. Use pattern name If patterns are loaded from a file, use names of pattern sequences as annotation names. The name from the parameters is used by default. Max Mismatches Maximum number of mismatches between a substring and a pattern. Allow Insertions/Deletions Takes into account possibility of insertions/deletions when searching. By default substitutions are only considered. Search in Translation
Translates a supplied nucleotide sequence to protein and searches in the translated sequence. Support ambiguous bases
Performs correct handling of ambiguous bases. When this option is activated insertions and deletions are not considered. Qualifier name Name of qualifier in result annotations which is containing a pattern name. Output data: On this page you can modify output parameters.