Some mutations could have a chromatogram signal lesser than the reference one. This situation very often appears if the mix of cells (healthy and oncological) has been being analyzed. That means that the mutation signal could be different (due to a different concentration of oncological cells in a sample).
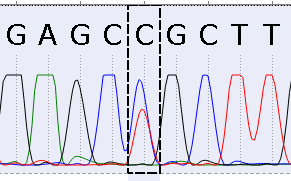
Look at the picture below. The signal of the "C" nucleotide (blue) is the highest. But there is a signal of the "T" nucleotide (red), which is high enough too. The second strongest signal is the thing we call Alternative mutation.
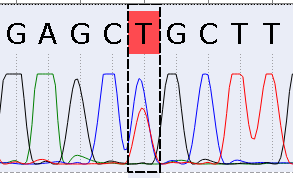
It's very important to make Alternative mutations clearly visible. To do this, you can use the function of the Alternative mutation. When the function is enabled and an alternative mutation fits settings, the corresponding character will be replaced with the alternative one. The replacement causes the difference between the read and the reference character, which means, that the read character will be highlighted:
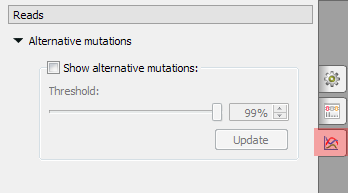
Alternative mutations settings are presented on the following picture:
Show alternative mutations - enable Alternative mutations and show them if checked, disable and hide otherwise.
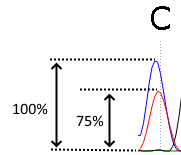
Threshold - how high the second strongest signal (corresponding to the firsts strongest signal) should be to be visible. Look at the picture below. The height of the second signal is 75% of the height of the first signal. That means, that the threshold value should be set to 75% (or below) to make this alternative mutation visible.
Update - click on this button to update the alignment corresponding to the set values.