This workflow performs multiple sequence alignment with MUSCLE algorithm and saves the resulting alignment to Stockholm document. Source data can be of any format containing sequences or alignments.
How to Use This Sample
If you haven't used the workflow samples in UGENE before, look at the "How to Use Sample Workflows" section of the documentation.
Workflow Sample Location
The workflow sample "Align Sequences with MUSCLE" can be found in the "Alignment" section of the Workflow Designer samples.
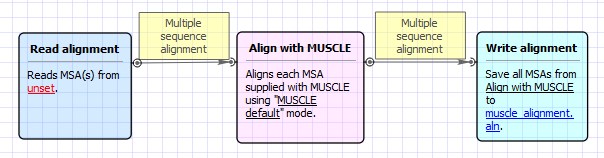
Workflow Image
The workflow looks as follows:
Workflow Wizard
The wizard has 2 pages.
Input MSA(s): On this page you must input multiple alignments file(s).
Align Sequences with MUSCLE: On this page you can modify MUSCLE and output parameters.
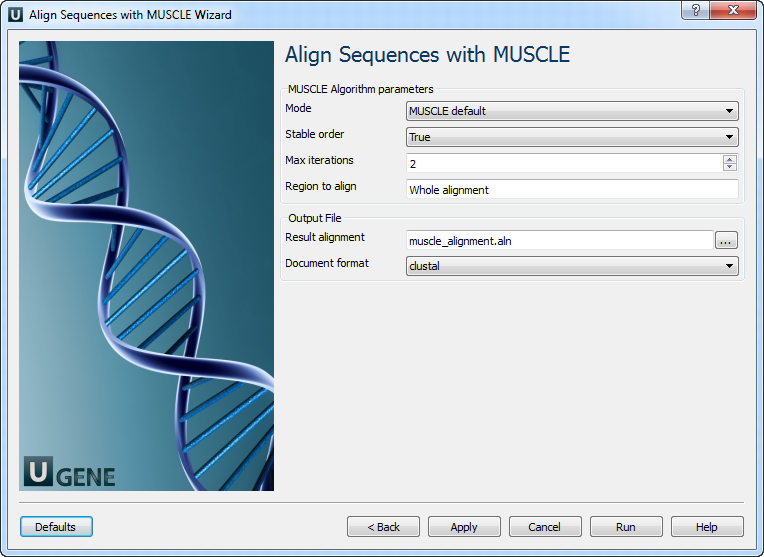
The following parameters are available:
Mode Selector of preset configurations, that give you the choice of optimizing accuracy, speed, or some compromise between the two. The default favors accuracy. Stable order Do not rearrange aligned sequences (-stable switch of MUSCLE).
Otherwise, MUSCLE re-arranges sequences so that similar sequences are adjacent in the output file. This makes the alignment easier to evaluate by eye.
Max iterations Maximum number of iterations. Region to align
Whole alignment or column range e.g. 1..100.
Result alignment
Location of output data file. If this attribute is set, slot "Location" in port will not be used.
Document format
Document format of output file.