Open a DNA molecule you want to cut into fragments.
Digestion into fragments is performed using restriction enzymes. So before continuing make sure that the restriction analysis has been performed. Refer chapter Restriction Analysis for details.
Select either the Tools ‣ Cloning ‣ Digest into Fragments item or the Actions ‣ Cloning ‣ Digest into Fragments item in the main menu or the Cloning ‣ Digest into Fragments item in the context menu.
The Digest Sequence into Fragments dialog appears:
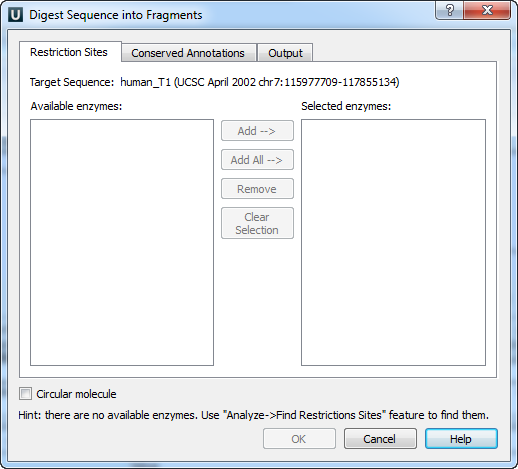
On the Restriction Sites tab of the dialog you can see the name of the molecule, the list of restriction enzymes found during the restriction analysis that can cut the molecule and the list of enzymes selected to perform the digestion.
To digest the sequence into fragments you should select at least one enzyme.
To move an enzyme to the Selected enzymes list click on it in the Available enzymes list and press the Add button. Note that you can select several items in a list by holding the Ctrl key while clicking on the items.
To select all available enzymes press the Add All button.
To remove enzymes from the Selected enzymes list select them in the list and press the Remove button.
To remove all items from the Selected enzymes list press the Clear Selection button.
On the Conserved Annotations tab of the dialog you can select the annotations that must not be disrupted during cloning.
On the Output tab of the dialog you can select the file to save the new molecule to.
As soon as the required parameters are selected press the OK button. The fragments will be saved as annotations.
Also all the generated fragments are available in the task report:
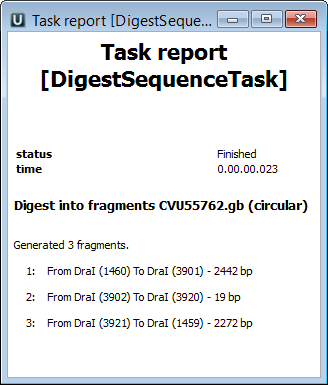
Refer to Notifications to learn more about task reports.