Originally, MAFFT is a multiple sequence alignment program for UNIX-like operating systems. However, currently, it is available for macOS, Linux, and Windows. It is used for both nucleotide and protein sequences.
MAFFT homepage: http://mafft.cbrc.jp/alignment/software
To make MAFFT available from UGENE:
Install the MAFFT program on your system.
Set the path to the MAFFT executable on the External tools tab of UGENE Application Settings dialog.
For example, on Windows, you need to specify the path to the mafft.bat file.
To use MAFFT open a multiple sequence alignment file and select the Align with MAFFT item in the context menu or in the Actions main menu. The following dialog appears:
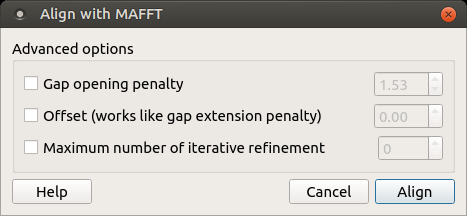
The following parameters are available:
Gap opening penalty — Gap opening penalty at group-to-group alignment.
Offset (works like gap extension penalty) — offset value, which works like the gap extension penalty, for group-to-group alignment.
Maximum number of iterative refine — specifies the number of cycles of iterative refinement to perform.