To find tandem repeats, select the Analyze ‣ Find tandems... context menu item in the Sequence View window.
In the opened dialog you can specify the tandem search parameters, the region to search in and the result parameters:
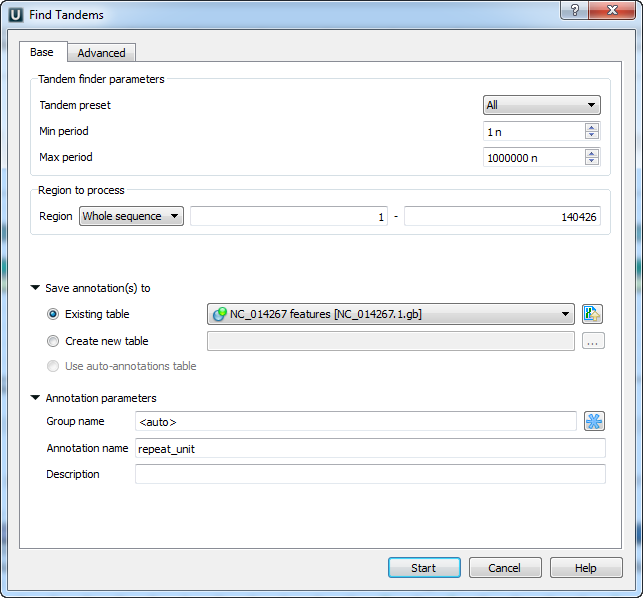
The dialog parameters:
Tandem preset — specify the tandem repeats parameters with predefined values by selecting the available preset:
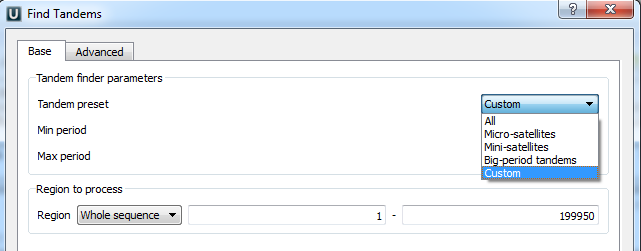
Min period, Max period — the minimum and maximum acceptable repeat length measured in base symbols.
Region to process — specify the region to search in the whole sequence, a custom region or the region of the current selection (if any).
In the Save annotation(s) to group you can set up a file to store annotations. It could be either an existing annotation table object, a new annotation table or auto-annotations table (if it is available).
In the Annotation parameters group you can specify the name of the group and the name of the annotation. If the group name is set to <auto> UGENE will use the group name as the name for the group. You can use the ‘/’ characters in this field as a group name separator to create subgroups. If the annotation name is set to by type UGENE will use the annotation type from the Annotation type: table as the name for the annotation. Also you can add a description in the corresponding text field.
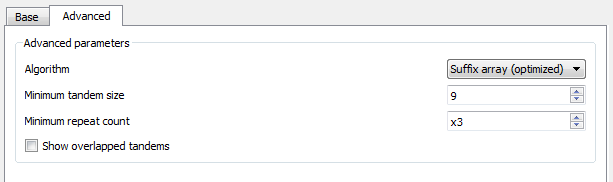
Additional search options can be found in the Advanced tab:
Algorithm — the algorithm parameter allows to select the search algorithm. The default and a fast one is optimized suffix array algorithm.
Minimum tandem size — the minimum tandem size sets the limit on minimum acceptable length of the tandem, i.e. the minimum total repeats length of the searched tandem.
Minimum repeat count — the minimum number of repeats of a searched tandem.
Show overlapped tandems — check if the plugin should search for the overlapped tandems, otherwise keep unchecked.