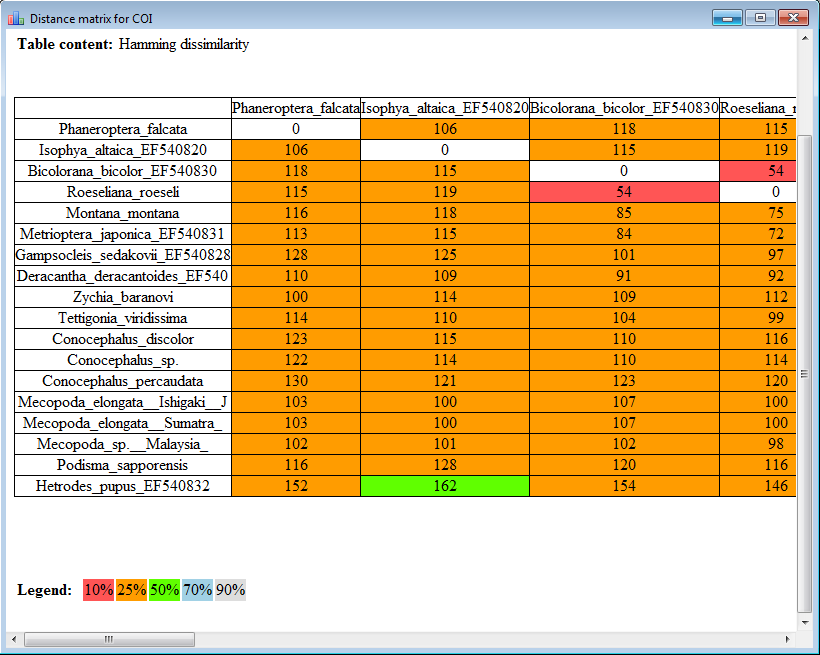
Using the Alignment Editor you can also create a distance matrix of a multiple sequence alignment.
To create a distance matrix, use the Statistics ‣ Generate distance matrix item in the Actions main menu or in the context menu.
The dialog will appear:
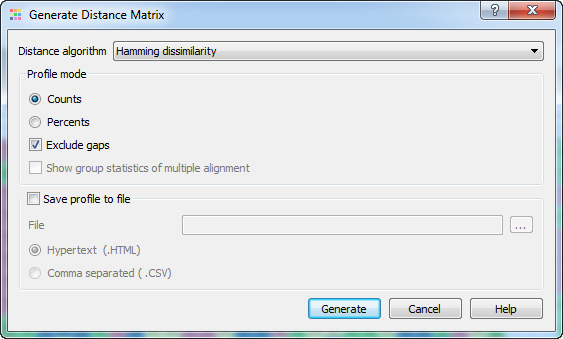
The following parameters are available:
Distance algorithm - there are two distance algorithms: "Hamming distance" for dissimilarity and "Simple similarity" for similarity.
Profile mode: Counts/Percents — select the Percents to have scores shown as percents in the report. Also you can Exclude gaps.
Show group statistics of multiple alignment - shows group statistics when the collapsing is switched on.
Save profile to file — allows to save profile to a file in the HTML or CSV format. The CSV format is convenient for further processing in worksheets editors like Excel.
The result profile in the HTML mode: