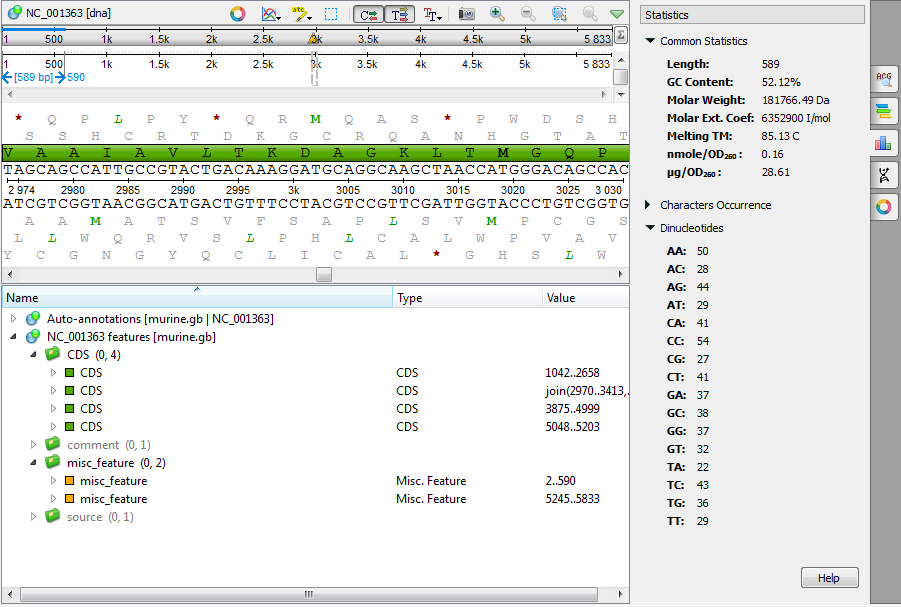
Context information about a sequence can be found on the Statistics tab in the Options Panel. All information is contextual, i.e. it shows statistics about the currently selected region (on the selected sequence). The tab includes information about:
- Common statistics
- Length - number of bases in the analyzed sequence
- GC content - the molar percentage of guanine and cytosine bases in an oligonucleotide sequence
- Molar weight - is the sum of the atomic masses of the constituent atoms for 1 mole of oligonucleotide
- Molar ext. coefficient - the molar extinction coefficient is a physical constant that is unique for each sequence and describes the amount of absorbance at 260nm (A260) of 1 mole/L DNA solution measured in 1 cm path-length cuvette
- Melting TM - melting temperature is the temperature at which an oligonucleotide duplex is 50% in single-stranded form and 50% in double-stranded form
- nmole/OD260 - the amount of oligonucleotide in nanomoles that, when dissolved in 1 mL volume, results in 1 unit of absorbance at 260 nm with a standard 1 cm path-length cuvette
- μg/OD260 - the amount of oligonucleotide in micrograms that, when dissolved in 1 mL volume, results in 1 unit of absorbance at 260 nm with a standard 1 cm path-length cuvette
- Characters occurrence
- Dinucleotides occurrence (for sequences with the standard DNA and RNA alphabets)
To copy the statistical information about a sequence select it on the Options Panel and choose the copy item in the context menu, or use the Ctrl+C shortcut.