To search transcription factor binding sites in a DNA sequence select the Analyze ‣ Search TFBS with SITECON... context menu item.
In the appeared search dialog you must select a file with TFBS profile. The profiles supplied with UGENE are placed in the $UGENE/data/sitecon_models folder.
After the profile is loaded the threshold-filter is populated with values read from profile. You can use the filter to remove low-scoring regions from the result.
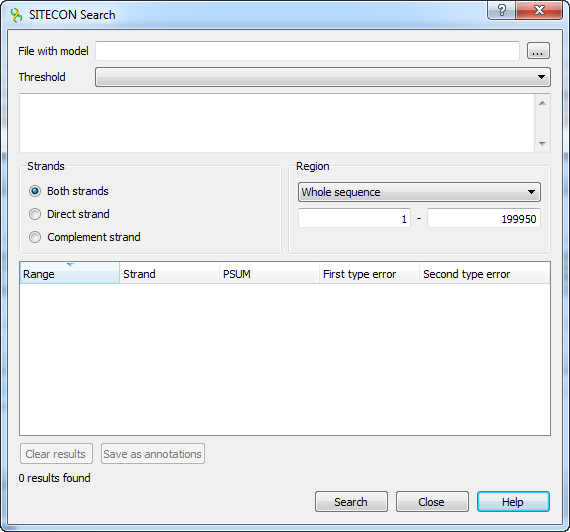
The regions found by SITECON algorithm can be saved as annotations to the DNA sequence in the Genbank format.
Every SITECON profile supplied with UGENE contains complete information about calibration settings provided to UGENE team by the author of SITECON.
The original TFBS alignments used to calculate profiles can be requested directly from the author of SITECON.