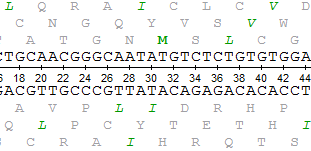
When a nucleotide sequence is opened in the Sequence View, the sequence and its complementary sequence are shown by default in the Details View.
Showing/hiding the amino acid sequences
It is possible to translate them and show also the corresponding amino acid sequences:
The translation settings are available on the left toolbar of the Details View:

The Show/hide amino acid translations menu allows one to set up the mode of the amino acid sequences visualization:
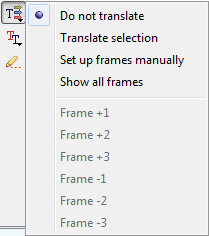
The following options are available:
- Do not translate— hide the amino acid sequences.
- Translate selection — translate only a selected region of the sequence and the complementary sequence.
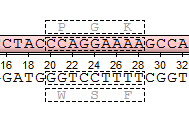
- Set up frames manually — select the reading frames to show. There are three frames for the sequence ("+1", "+2", "+3") and three frames for the complementary sequence ("-1", "-2", "-3"). Note that the complementary frame items are hidden in the menu, if the complementary sequence is hidden.
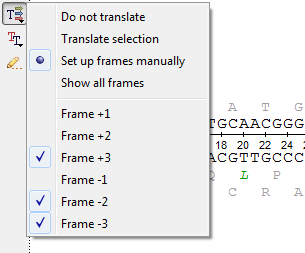
- Show all frames — show all amino acid sequences.
Setting the genetic code
The default value for the genetic code for a nucleotide sequence translation is read by UGENE from the sequence file when it is available. One can also set up the genetic code for the sequence using the Select genetic code menu:
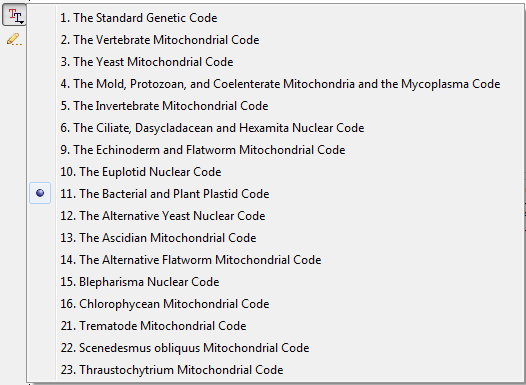
All analysis routines (like HMMER, ORF finding, etc.) will use this code by default.
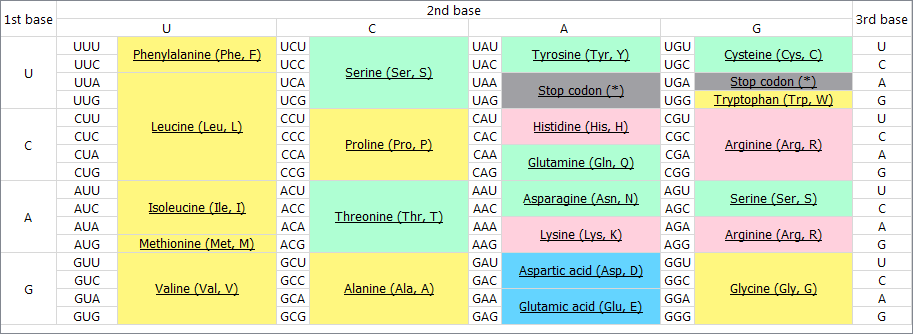
Codon table
To refresh the knowledge about the amino acid codes, select the Show codon table button on the Sequence View global toolbar. The codon table appears at the upper part of the window: