This feature allows to search for primer pairs that span introns on the genomic sequence or exon junctions on the mRNA sequence.
Note that RT-PCR design is only available for mRNA/cDNA sequences with annotated exons. There are several ways to obtain the cDNA for a corresponding DNA sequence.
- From NCBI or ENSEMBL database.
For example, one can download the TMPRSS2 transcript variant 1 from NCBI Genbank using identifier NM_001135099.1.
This can be also done from UGENE using option Access remote database or Search NCBI Genbank. - Align the genomic and cDNA sequences using spliced aligner.
For this option one must have both genomic and cDNA sequences.
In UGENE the spliced alignment can be performed using the Spidey tool.
To run the alignment open the genomic sequence and select action Align ‣ Align to mRNA sequence.
The generated exon annotations can be then exported using action Export ‣ Export sequence of selected annotations
To design primers for your mRNA sequence and go to the RT-PCR tab of the Primer Designer dilaog:
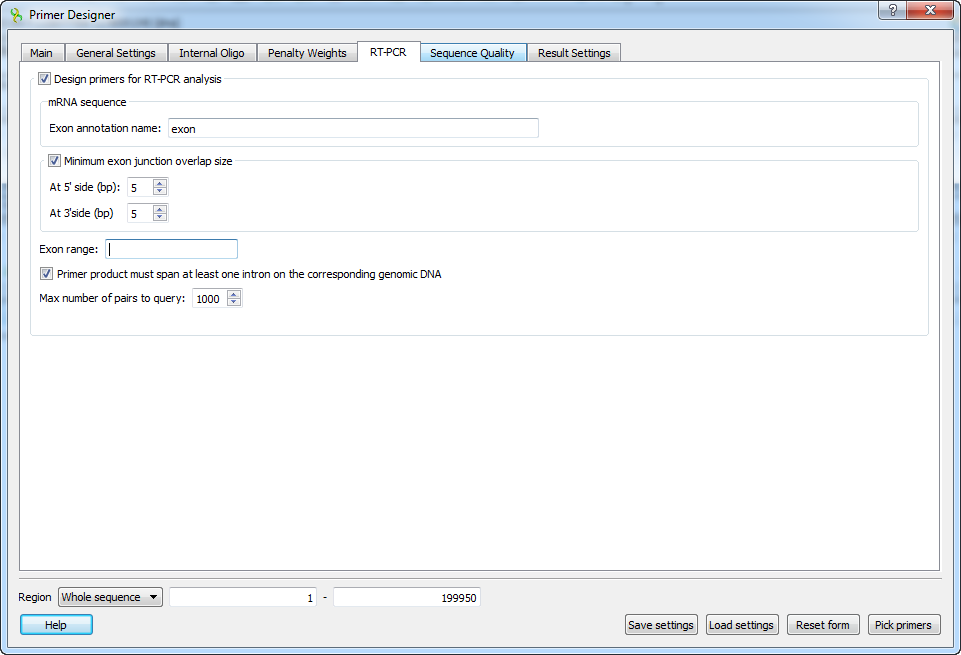
The following parameters are available:
Exon annotation name
To detect exon boundaries UGENE searches for exonic annotations. This option allows to set custom name for annotations denoting exons. Default value is "exon"
Minimum exon junction overlap size
If checked, then only the pairs with at least one of the primers overlapping exon junction in the mRNA sequence will be selected.
At 5' side (bp)
Minimum overlap size on the 5' side of the exon junction. Default is 5 bp.
At 3' side (bp)
Minimum overlap size on the 3' side of the exon junction. Default is 5 bp.
Exon range
This option allows to limit the sequence region, where the primers are searched for. For example, setting value "3-5" will limit the search to a sequence region consisting of exons 3,4,5 of the transcript, as defined by the order in the sequence. Default value is an empty string, which means that there are no limitations.
Span at least one intron
This option makes sure that primer product should span an intron on the genomic sequence i.e the forward and reverse primers must be located in different exons. The option is enabled by default.
Max numbers of pairs to query
The algorithm applied in RT-PCR primer design first searches for all available primers in a given sequence. Then it filters the detected pairs to make sure that they satisfy the selected configuration. This option allows to set the maximum number of pairs for the initial search query. Larger number will result in increased sensitivity, but also in a longer running time. Default value is 1000.
Important: using the RT-PCR primer design tab will reset the values set in the Exlcuded regions and Targets of the Main configuration tab. Additionally if the Exon range option is set, the defined sequence region will be ignored.