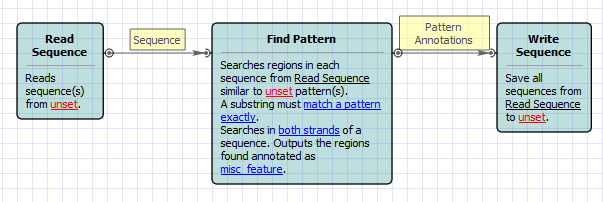
This simple workflow finds patterns in you sequences and save them as annotations. You can use the workflow to map primers, regulatory signals, genes, etc. It loads any set of sequences from your files or folders and finds patterns in them. Just specify a dataset for the algorithm in the "Read sequence" element. Patterns are entered in comma-delimited format in the corresponding field of the "Find Pattern" element. Also you can load patterns from a file. In that case names of patterns can be saved as names of annotations. Files with patterns can be in any sequence format or in newline-delimited format.
How to Use This Sample
If you haven't used the workflow samples in UGENE before, look at the "How to Use Sample Workflows" section of the documentation.