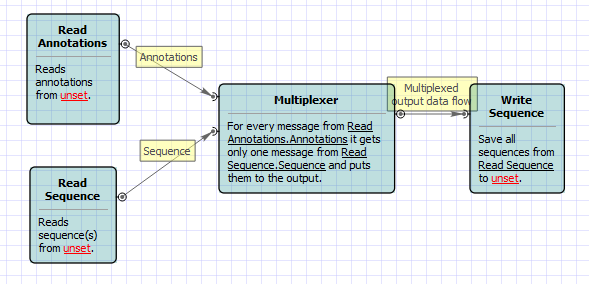
If you have a list of files with sequences and separate files with annotation and you want to merge sequences and annotation, this workflow might help you. For instance, you have sequence in FASTA format and separate annotation in GFF. You want to merge them and write annotated sequences into Genbank files. Be default, multiplexer takes sequences and annotation one by one, sticks one annotation to one sequence and passes it to the output. But you may change that behavior in parameters of Multiplexer.
How to Use This Sample
If you haven't used the workflow samples in UGENE before, look at the "How to Use Sample Workflows" section of the documentation.