Suppose you have genomes and you want to characterize them. One of the ways to do that is to build a table of what genes are in each genome and what are not there.
- Create a local BLAST db of your genome sequence/contigs. One db per one genome.
- Create a file with sequences of genes you what to explore. This file will be the input file for the workflow.
- Setup location and name of BLAST db you created for the first genome.
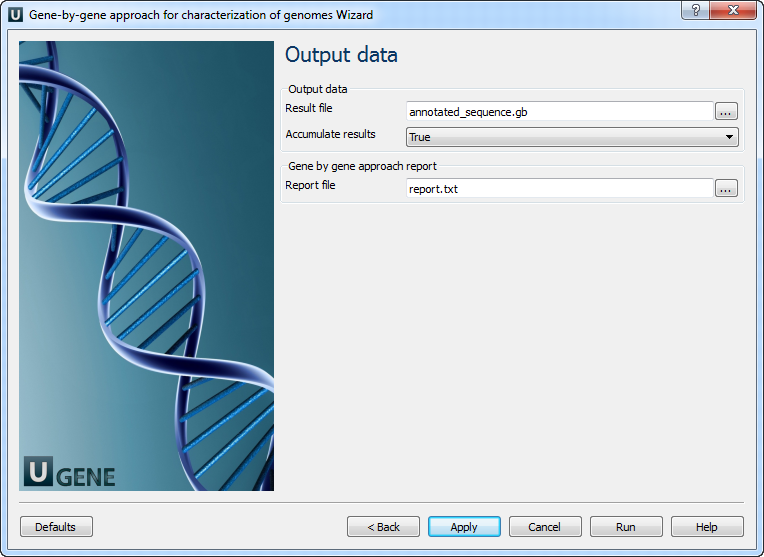
- Setup output files: report location and output file with annotated (with BLAST) sequence. You might want to delete the "Write Sequence" element if you do not need output sequences.
- Run the workflow.
- Run the workflow on the same input and output files changing BLAST db for each genome that you have.
As the result you will get the report file. With "Yes" and "No" field. "Yes" answer means that the gene is in the genome. "No" answer MIGHT mean that there is no gene in the genome. It is a good idea to analyze all the "No" sequences using annotated files. Just open a file and find a sequence with a name of a gene that has "No" result.
How to Use This Sample
If you haven't used the workflow samples in UGENE before, look at the "How to Use Sample Workflows" section of the documentation.
Workflow Sample Location
The workflow sample "Gene-by-gene Approach for Characterization of Genomes" can be found in the "Scenarios" section of the Workflow Designer samples.
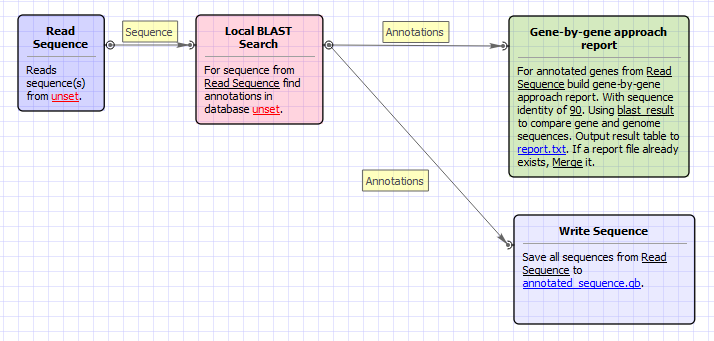
Workflow Image
The workflow looks as follows:
Workflow Wizard
The wizard has 3 pages.
Input sequence(s): On this page you must input sequence(s).
BLAST search: On this page you can modify BLAST search parameters.
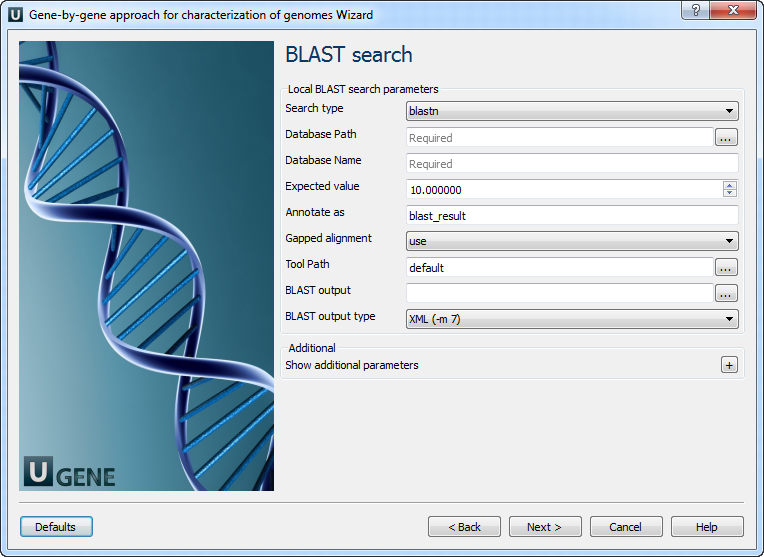
The following parameters are available:
Search type Select type of BLAST searches. Database Path Path with database files. Database Name Base name for BLAST DB files. Expected value This setting specifies the statistical significance threshold for reporting matches against database sequences. Annotate as Name for annotations. Gapped alignment
Perform gapped alignment. Tool Path
External tool path. BLAST output Location of BLAST output file. BLAST output type Type of BLAST output file. Temporary directory Directory for temporary files. Gap costs Cost to create and extend a gap in an alignment. Match scores Reward and penalty for matching and mismatching bases. Output data: On this page you can modify output parameters.