The DNA Annotator plugin Find Group of Annotated Regions feature provides an algorithm to search for sequence regions that contain a predefined set of annotations.
Usage example:
Open the Sequence View for a sequence that has annotations. A good candidate here Open a DNA sequence in the Sequence View. There are two ways to open the Find Repeats dialog:
- By clicking on the toolbar button:
- By selecting the Analyze ‣ Find annotated regions... context menu item:
This tool has been designed to search for annotations that intersect (or completely overlap - it depends on the specified parameters) other, already existing annotations of a given sequence. Let's look at the example:
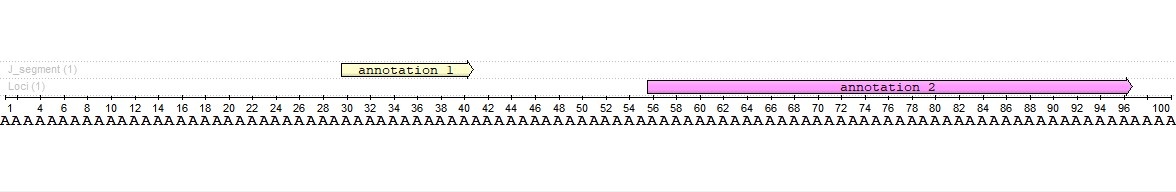
We have a sequence with two annotations. Annotations have different lengths and do not intersect each other. The annotation 2 length is four times the annotation 1 length (41 vs. 11 bases).
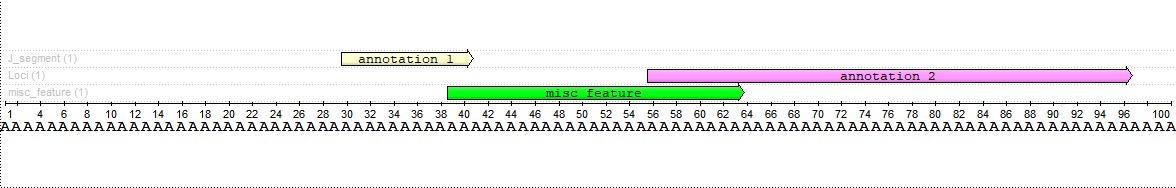
Using this function, we can find an annotation that intersects both source annotations and captures their shares depending on their lengths. For example, lets find an intersection 25 bases long. We will have the following annotation:
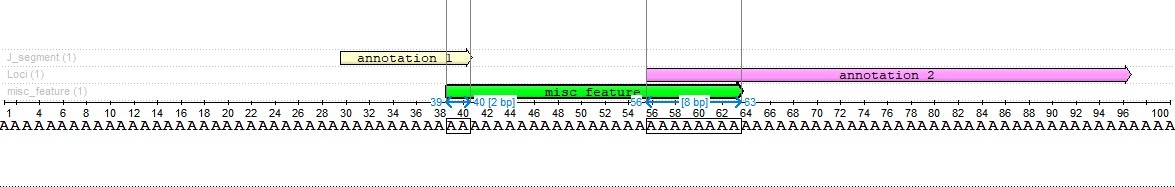
As we can see, the intersection with the first annotation is two characters long, and the intersection with the second annotation is eight characters long. This result was chosen because the second annotation is four times the length of the first annotation.
NOTE: A good candidate for this feature could be any file in Genbank format with a rich set of annotations.
Select the Analyze ‣ Find annotated regions item in the context menu. The dialog will appear:
| HTML |
|---|
<center>
<br>
<img src="/wiki/download/attachments/4227609/DNA Annotator.png"/>
<br>
</center> |
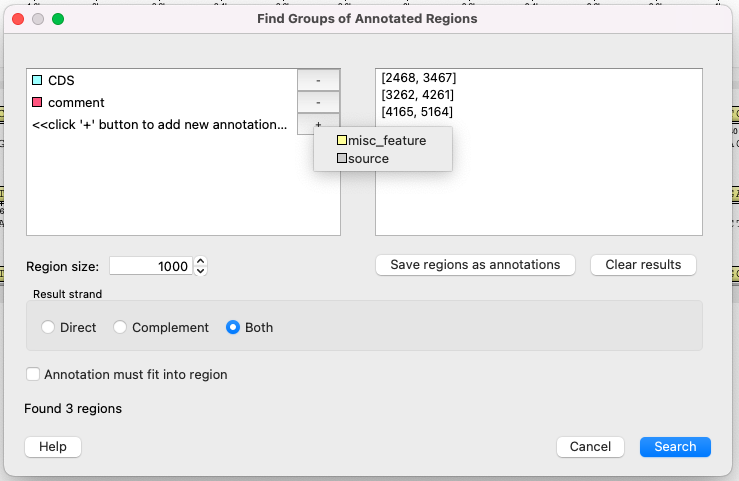
Using this dialog you can search for DNA sequence regions that contain every annotation from the list on the left side. The found regions are displayed on the right side of the dialog.
Use the Save regions as annotations button to store the regions as new annotations to the sequenceFASTA is not the best option, because this format does not store annotations.
Parameters
The following parameters are avaliable:
- Left window - annotations to search intersection regions for.
- Right window - the list of possible intersection regions.
- Region size - the length of the new intersection region.
- Result strand - select the DNA strand whose annotations will be considered in the search. If, for example, the "Complement" strand is selected, but all choosen annotations are on the direct strand, than nothing will be found.
- Annotation must fit into region - all annotations, choosen at the left window, must fit completely to the result annotation (completely - and not just in a few characters).
- Save regions as annotations - store results to annotations.
- Clear results - clear the result table.