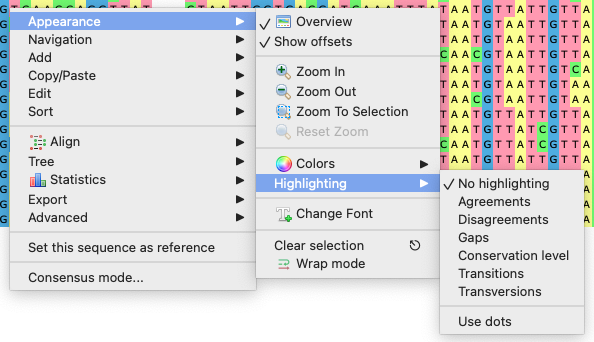
To apply an alignment highlighting mode, select it in the Highlighting context menu:
...
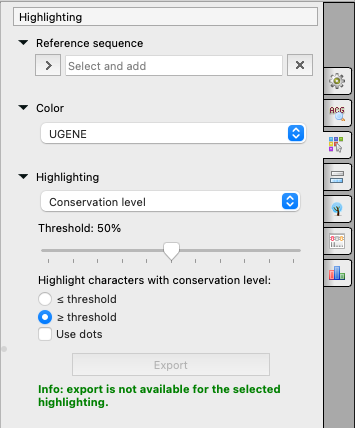
or on the Highlighting tab of the Options Panel:
| HTML |
|---|
<center>
<br>
<img src="/wiki/download/attachments/16122233/Highlighting Alignment_1.png"/>
<br>
</center> |
The
Referense sequence
Some highlighting algorithms consider not the alignment itself, but changes of the whole alignment (minus one sequence) from one sequence. This one sequence is called the reference sequence and you may set it here.
NOTE: not all highlighting algorithms take the reference into account, those that will be marked RS below.
Color
Different color schemes. Pay attention, that DNA and AMINO alignment have different color schemes:
- No colors - no colors at all.
The list of Jalview compatible color schemes for amino acid alphabet only:
- Buried Index,
- Turn propensity,
- Strand propensity,
- Helix propensity,
- Hydrophobicity,
- Tailor,
- Zappo.
Look at the following table for the color details:
The list of DNA/RNA/amino alphabet color schemes:
- Percentage identity - the higher the frequency percentage value of the base in the certain column, the bluer the corresponding base (only the highest frequency base is colored),
- Percentage identity (gray) - the same as Percentage identity, but in black and white,
- UGENE (default) - classic UGENE color scheme,
- UGENE Sanger - UGENE color scheme, which is mainly used in the Sanger Reads editor,
- Weak similarities - look here for details.
The list of DNA/RNA alphabet color schemes:
- Jalview - Jalview compatible color scheme,
- Percentage identity (colored) - look here for details.
Highlighting
Choose, what types of bases will be colored with the color scheme choosen. The following modes are available:
Agreements (RS) — highlights symbols that coincide with the reference sequence.
DisagreementsDisagreements (RS) — highlights nucleotides symbols that differ from the reference sequence.
- Gaps - highlights gaps only.
- Conservation level - highlights conservation level of symbols in a multiple alignment >= or <= treshhold. To select the conservation parameters use the Highlighting Options Panel tab.
- Transitions - highlights transitions.
- Transversions - highlights transversions.
- highlights bases, which frequency percentage value of the column is greater/lower, than the corresponding threshold.
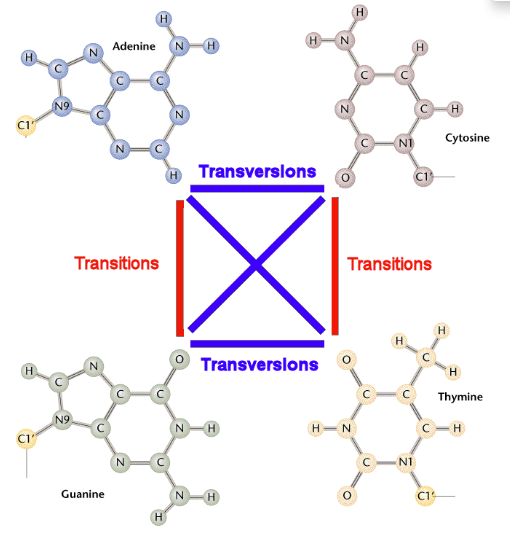
- Transitions(RS) - highlights transitions of the referense base. Transitions are interchanges of two-ring purines (A <-> G), or of one-ring pyrimidines (C <-> T): they therefore involve bases of similar shape.
- Transversions(RS) - highlights transversions of the referense base. Transversions are interchanges of purine for pyrimidine bases, which therefore involve exchange of one-ring & two-ring structures.
Look at the following pictures for details about transitions and transversions:
To use dots instead of symbols which are not highlighted check the Use dots checkbox in the Options Panel or use the Highlighting->Use dots context menu item.To select a reference sequence use the Set this sequence as reference context menu or Reference sequence field in the Highlighting tab of the Options Panel..
Export highlighted
Also you can export highlighting with a help of the Export button in the Options Panel or by the Export->Export highlighted context menu item. The following dialog will appear:
| HTML |
|---|
<center>
<br>
<img src="/wiki/download/attachments/7667818/Highlighting Alignment_2.png"/>
<br>
</center> |
Select file to export, exported area and click on the Export button. The task report will appear in the NotificationsCheck the corresponding page fpr details.