In silico PCR used to calculate theoretical polymerase chain reaction (PCR) results using a given set of primers (probes) to amplify DNA sequences.
In silico PCR is available only for nucleic sequences. To use it in UGENE open a DNA sequence and go to the In Silico PCR tab of the Options Panel:
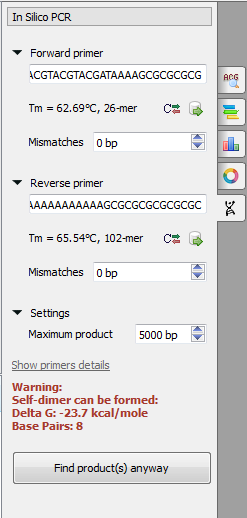
Here the following parameters are available:
Forward primer - forward primer.
Reverse primer - on the opposite strand from the forward primer.
Mismatches - mismatches limit.
Maximum product - maximum size of amplified region.
Type two primers for running In Silico PCR. Note, if the primers pair is invalid for running the PCR process then the warning is shown. For making a primer sequence reverse-complement click the Reverse-complement button:
Also, primers can be chosen from a primers library. Click Show primers details for seeing statistic details about primers.
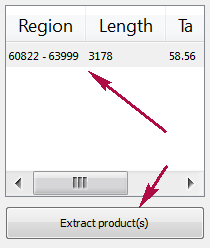
When you run the process, the predicted PCR products appear in the products table. There are three columns in the table: region of product in the sequence, product length and preferred annealing temperature. Click the product for navigating to its region in the sequence. Click the Extract product(s) button for exporting a product(s) in a file. Or use double click for that.