UGENE integrates a set of tools for taxonomy classification of microorganisms using whole-genome shotgun sequencing data. The tools are Kraken, CLARK, DIAMOND, etc. See, for example, "Parallel NGS Reads Classification" sample workflow that allows one to classify input FASTQ files with these tools working in parallel and then join their output with another tool WEVOTE.
The tools are available on 64-bit macOS or Linux operating systems only. Also, as the tools are quite resource-consuming, it is recommended to have at least 16 Gb of RAM available.
To use these tools one should provide appropriate taxonomy data and reference data, specific for a tool. Some reference databases are provided for each of the tool. One can use these data or build a custom database (see, for example, workflow element "Build Kraken Database").
It is recommended to use the UGENE Online Installer package to install and automatically configure the data. However, if the Internet is not available on the target computer, or it is required to use another UGENE package for some other reason, follow the instructions below on how to download and configure the data.
Download NGS classification data
Use links in the "Data for NGS taxonomy classification" section on the "Download UGENE and components" page to download the data.
Make sure to have enough disk space on the target computer.
See the list of the available downloads in the table below.
| Data | Archive size | Unpacked data size | Description | Data source |
|---|---|---|---|---|
| NCBI taxonomy classification | 2.5 Gb | 31 Gb | This includes a set of taxonomy data files from NCBI. These data should be present for any type the NGS classification analysis. | Original data were downloaded from the NCBI FTP (ftp://ftp.ncbi.nlm.nih.gov/pub/taxonomy/). |
| NCBI RefSeq bacterial genomes | 130 Gb | 132 Gb | The data can be used to build a database for CLARK-l (light version of CLARK), CLARK, or Kraken. As UGENE integrates modified version of CLARK/CLARK-l, it is possible to provide *.gz archives as input for building the database. In particular, "CLARK-l DB: RefSeq bacterial+viral genomes" (see below) was generated using the archived data. Also, keep in mind that changing of some parameters of the "Classify Sequences with CLARK" element may cause re-building of the reference database. The reference data should be present in this case! For building a Kraken database usage of *.gz archives is not supported, it is required to unpack each *.gz file, so even more disk space will be required.
| Original data were downloaded from the NCBI FTP (ftp://ftp.ncbi.nlm.nih.gov/refseq/release/bacteria/bacteria*.genomic.fna.gz) |
| NCBI RefSeq viral genomes | 77 Mb | 77 Mb | Similarly to "NCBI RefSeq bacterial genomes", although the size of the data is rather small. The reference data are included into "CLARK-l DB: RefSeq bacterial+viral genomes" and "CLARK-l DB: RefSeq viral genomes" databases. | Original data were downloaded from the NCBI FTP (ftp://ftp.ncbi.nlm.nih.gov/refseq/release/viral/viral*.genomic.fna.gz). |
| NCBI RefSeq GRCh38 human genome | 837 Mb | 838 Mb | Similarly to "NCBI RefSeq bacterial genomes". The data are not included into any database, but provided in case one would like to use them when building a custom database. | Original data were downloaded from the NCBI FTP (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/CHR_*/hs_ref_GRC*chr*.fa.gz). |
| Kraken DB: MiniKraken 4Gb database | 2.5 Gb | 4.3 Gb | A sample reference database provided in UGENE for Kraken. It is a pre-built 4 GB database constructed from complete bacterial, archaeal, and viral genomes in RefSeq (as of Oct. 18, 2017). This can be used by users without the computational resources needed to build a Kraken database. However this contains only 2.7% of kmers from the original database. | Original data were downloaded using a link on the Kraken web site (https://ccb.jhu.edu/software/kraken/). |
| CLARK-l DB: RefSeq bacterial+viral genomes | 7.4 Gb | 11 Gb | One of the reference databases provided in UGENE for CLARK-l. The database was build using archived RefSeq bacterial and viral genomes. | See above. |
| CLARK-l DB: RefSeq viral genomes | 16 Mb | 72 Mb | One of the reference databases provided in UGENE for CLARK-l. The database was build using archived RefSeq viral genomes. | See above. |
| DIAMOND DB: UniRef50 | 5.2 Gb | 13 Gb | One of the reference databases provided in UGENE for DIAMOND. Note that unlike Kraken and CLARK, DIAMOND requires protein reference sequences as input. | Original data were downloaded from the Uniprot FTP (ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/uniref/uniref50/uniref50.fasta.gz). Then a DIAMOND database was built. |
| DIAMOND DB: UniRef90 | 13 Gb | 34 Gb | One of the reference databases provided in UGENE for DIAMOND. | Original data were downloaded from the Uniprot FTP (ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/uniref/uniref90/uniref90.fasta.gz). Then a DIAMOND database was built. |
| Total: | 161 Gb | 226 Gb |
Configure data
Data described above are stored as 7zip archives. After a file download, unpack it using an appropriate file archiver (for example, Keka on macOS).
The unpacked data are stored in a folders structure with the root folder called "data". For example, for "NCBI RefSeq viral genomes" the archive is called "ngs_classification.clark.viral_database.7z" and the unpacked data look as follows:
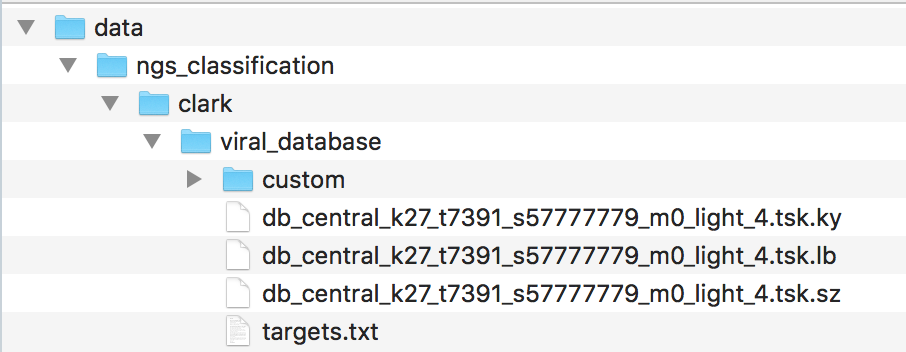
It is required to move these data to the UGENE data folder, following the hierarchical data structure:
- On Linux it is "data" folder, located in the UGENE installation folder.
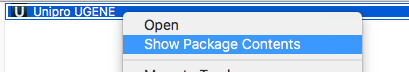
- On macOS the "data" folder is located inside the "Unipro UGENE.app" bundle. Right-click on the bundle, select "Show Package Contents", select "Contents -> MacOS -> data" folder.
Thus, all required data should be placed to the "ngs_classification" sub-folder of the UGENE data folder.