The Building Phylogenetic Tree dialog for the PhyML Maximum Likelihood method has the following view:
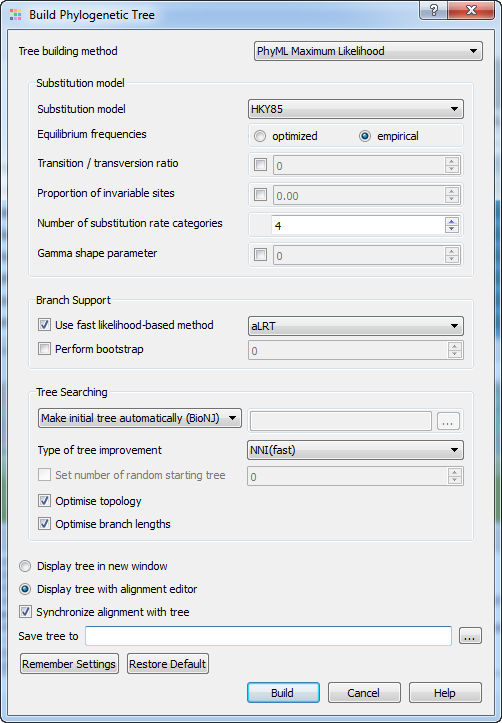
There following parameters are available:
Substitution model parameters - selection of the Markov model of substitution:
Substitution model - model of substitution.
Equilibrium frequencies - equilibrium frequencies.
Transition/transversion ratio - fix or estimate the transition/transversion ratio in the maximum likelihood framework.
Proportion of invariable sites - the proportion of invariable sites, i.e., the expected frequency of sites that do not evolve, can be fixed or estimated.
Number of substitution rate categories - number of substitution rate categories.
Gamma shape parameter - the shape of the gamma distribution determines the range of rate variation across sites.
Branch support parameters - selection of the method that is used to measure branch support:
Use fast likelihood method - use fast likelihood method.
Perform bootstrap - the support of the data for each internal branch of the phylogeny can be estimated using non-parametric bootstrap.
Tree searching parameters - selection of the tree topology searching algorithm:
Make initial tree automatically - initial tree automatically.
Type of tree improvement - type of tree improvement.
Set number of random starting tree - number of random starting tree.
Optimize topology - the tree topology is optimised in order to maximise the likelihood.
Optimize branch lengths - optimize branch lengths.
Display tree in new window - displays tree in new window.
Display tree with alignment editor - displays tree with alignment editor.
Synchronize alignment with tree - synchronize alignment and tree.
Save tree to - file to save the built tree.
Press the Build button to run the analysis with the parameters selected and build a consensus tree.