The Query Designer allows a molecular biologist to analyze a nucleotide sequence using different algorithms (Repeats finder, ORF finder, Weight matrix matching, etc.) at the same time imposing constraints on the positional relationship of the results obtained from the algorithms.
A user-friendly interface is used to create a schema of the algorithms and constraints.
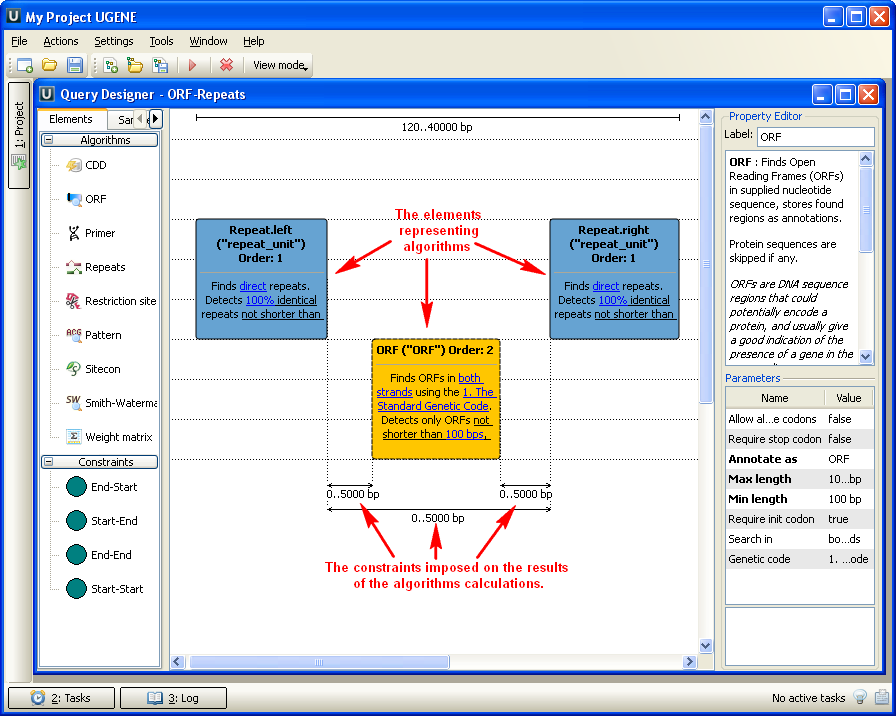
Alternatively, you can create / edit a schema using a text editor.
When the schema has been created and all its parameters have been set you can run it for a nucleotide sequence. The results are saved as a set of annotations to the specified file in the Genbank format. Also when you have query designer scheme you can analyze a nucleotide sequence from the sequence view with a help of this schema. Call the Analyze->Analyze with query schema context menu item for this.
To learn more about the Query Designer read the Query Designer Manual (follow the link on the UGENE documentation page)