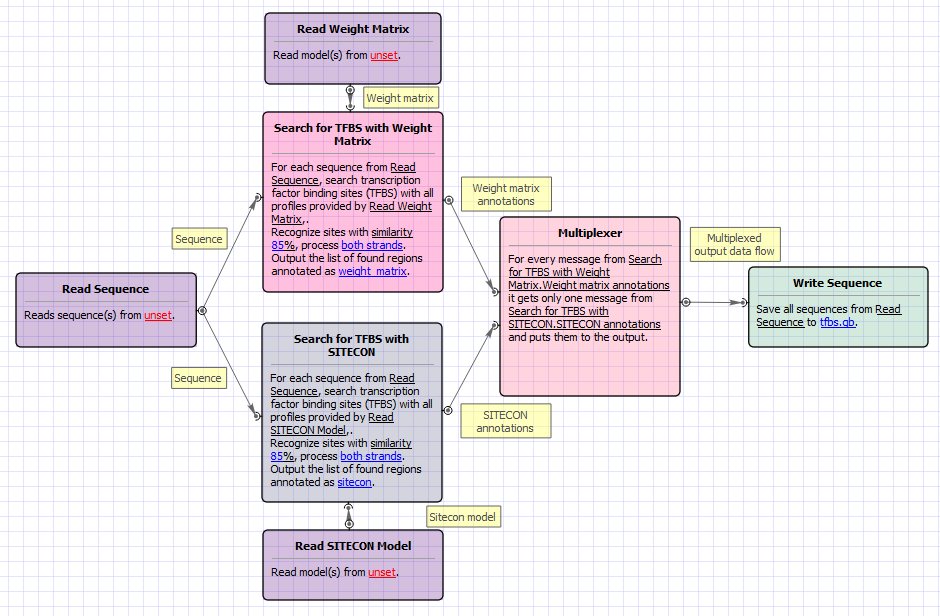
This sample shows how to search for transcription factor binding sites (TFBS) using two different approaches - weight matrices and SITECON models - and write the found TFBS annotations into one output file.
The workflow steps are these:
- The workflow reads the input sequences.
- Each sequence goes to the TFBS searching elements.
- Read Weight Matrix reads the input weight matrices. Read SITECON Model reads the input SITECON models. The data are also transferred to the TFBS searching elements.
- Each TFBS searching element produces the corresponding annotations.
- After that the two annotation data flows are multiplexed into one data flow.
- The multiplexed data and are written to the output file ("merged.gb", by default).
How to Use This Sample
If you haven't used the workflow samples in UGENE before, look at the "How to Use Sample Workflows" section of the documentation.
Workflow Sample Location
The workflow sample "Search for TFBS" can be found in the "Data Merging" section of the Workflow Designer samples.
Workflow Image
The workflow looks as follows:
Workflow Wizard
The wizard has 3 pages.
Input sequence(s): On this page you must input sequence(s).
Search for TFBS parameters: On this page you can modify search for TFBS parameters.
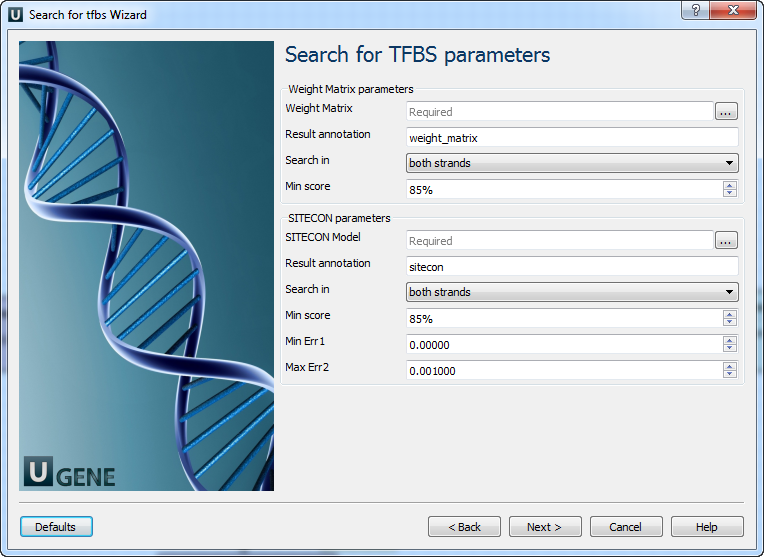
The following parameters are available:
Weight Matrix Semicolon-separated list of paths to the input files. Result annotation Annotation name for marking found regions. Search in Which strands should be searched: direct, complement or both. Min score
Minimum score to detect transcription factor binding site SITECON model
Semicolon-separated list of paths to the input files. Result annotation
Annotation name for marking found regions. Search in
Which strands should be searched: direct, complement or both. Min score
Minimum score to detect transcription factor binding site Min err1 Alternative setting for filtering results, minimal value of Error type I.
Note that all thresholds (by score, by err1 and by err2) are applied when filtering results.Max err2 Alternative setting for filtering results, max value of Error type II.
Note that all thresholds (by score, by err1 and by err2) are applied when filtering results.Output data: On this page you can modify output parameters.
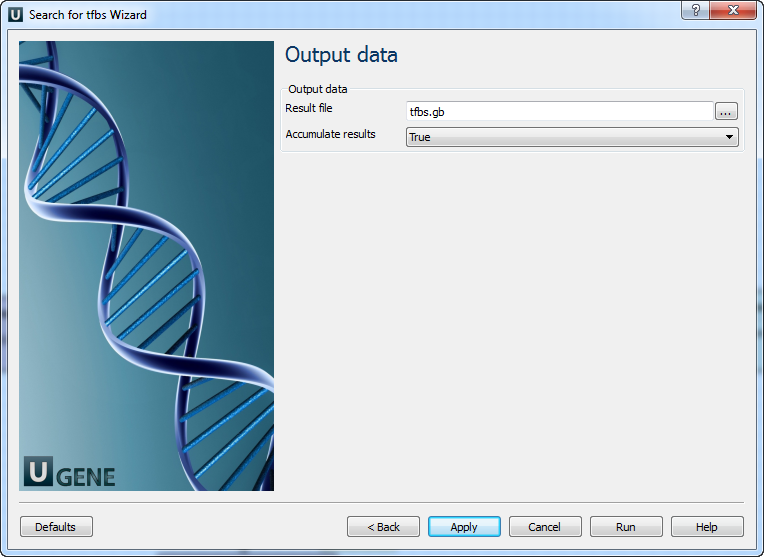
The following parameters are available:
Result file
Location of output data file. If this attribute is set, slot "Location" in port will not be used.
Accumulate results
Accumulate all incoming data in one file or create separate files for each input.In the latter case, an incremental numerical suffix is added to the file name.