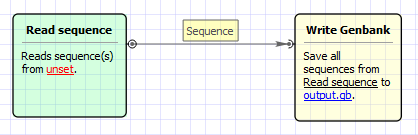
This workflow converts sequence file(s) of any format (including PDB, aligments etc) to Genbank document(s). If source format supports annotations, they are also saved as feature tables in target file. Sequence meta-information (accessions etc) is preserved as well.
How to Use This Sample
If you haven't used the workflow samples in UGENE before, look at the "How to Use Sample Workflows" section of the documentation.
Workflow Sample Location
The workflow sample "Convert Sequence to Genbank" can be found in the "Conversions" section of the Workflow Designer samples.
Workflow Image
The workflow looks as follows:
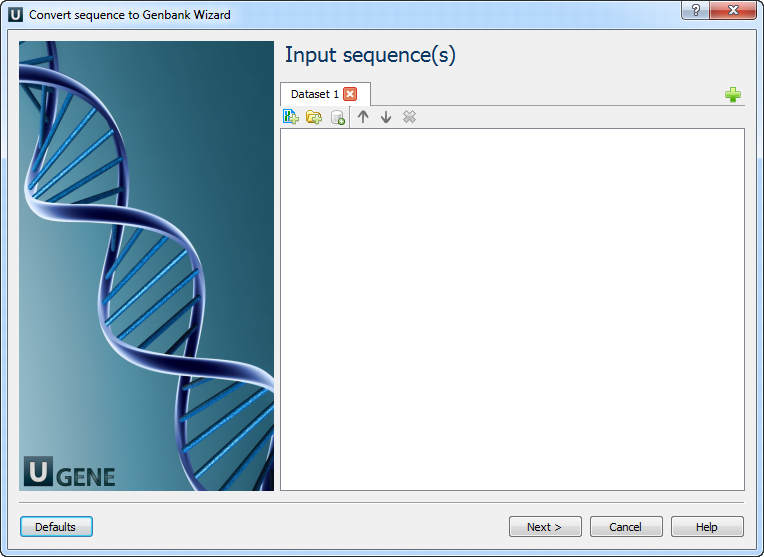
Workflow Wizard
The wizard has 2 pages.
Input sequence(s): On this page you must input sequence(s).
Output data: On this page you can modify output settings.
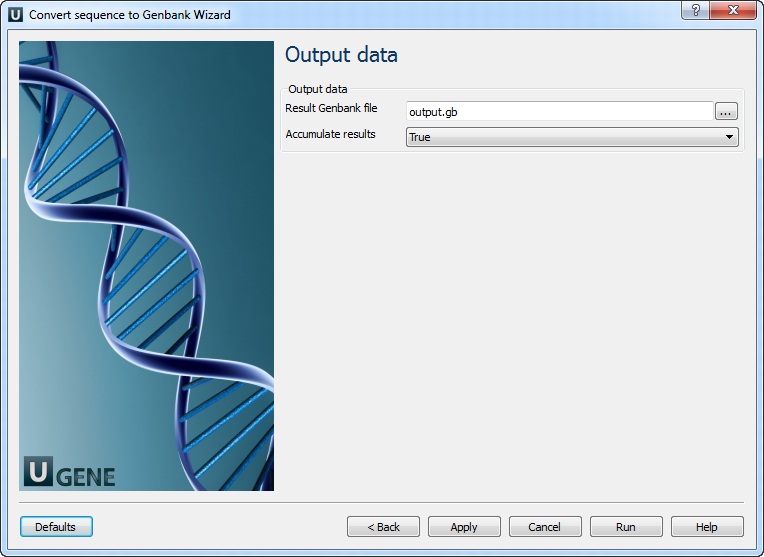
The following parameters are available:
Result Genbank file Location of output data file. If this attribute is set, slot "Location" in port will not be used. Accumulate results Accumulate all incoming data in one file or create separate files for each input.In the latter case, an incremental numerical suffix is added to the file name.