Use this workflow sample to extract unmapped reads from an input SAM/BAM file.
How to Use This Sample
If you haven't used the workflow samples in UGENE before, look at the "How to Use Sample Workflows" section of the documentation.
Workflow Sample Location
The workflow sample "Get Unmapped Reads" can be found in the "NGS" section of the Workflow Designer samples.
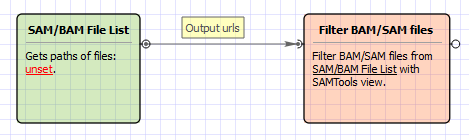
Workflow Image
The workflow looks as follows:
Workflow Wizard
The wizard has 3 page.
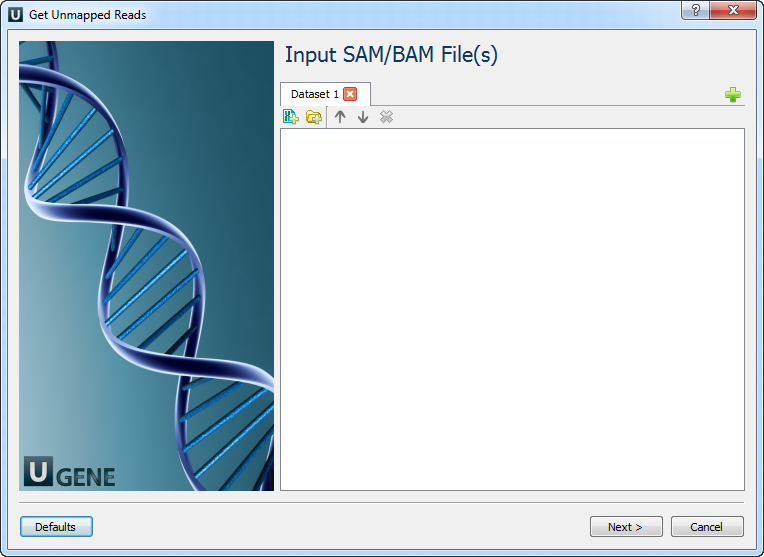
Input SAM/BAM File(s): On this page you need input SAM/BAM file(s).
Filtration: On this page you can change the filtration parameters.
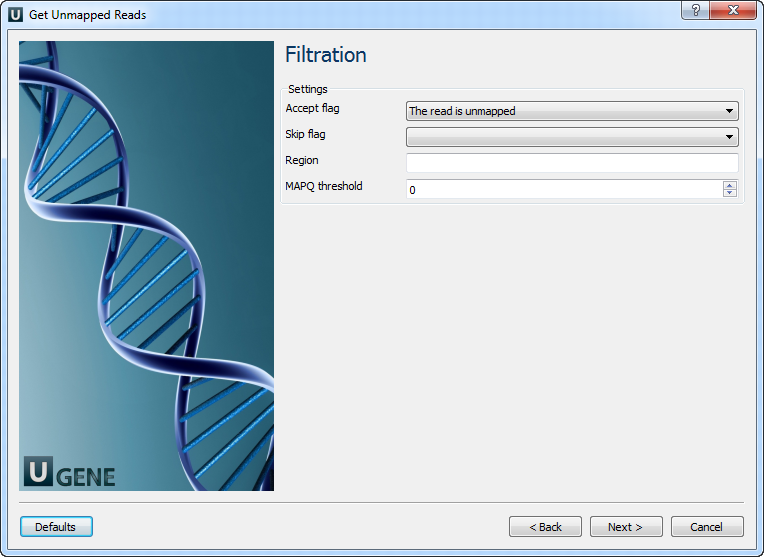
The following parameters are available:
Accept flag Only output alignments with the selected items. Select the items in the combobox to configure bit flag. Do not select the items to avoid filtration by this parameter. Skip flag Skip alignment with the selected items. Select the items in the combobox to configure bit flag. Do not select the items to avoid filtration by this parameter. Region Regions to filter. For BAM output only. chr2 to output the whole chr2. chr2:1000 to output regions of chr 2 starting from 1000. chr2:1000-2000 to ouput regions of chr2 between 1000 and 2000 including the end point. To input multiple regions use the space seprator (e.g. chr1 chr2 chr3:1000-2000). MAPQ threshold
Minimum MAPQ quality score.
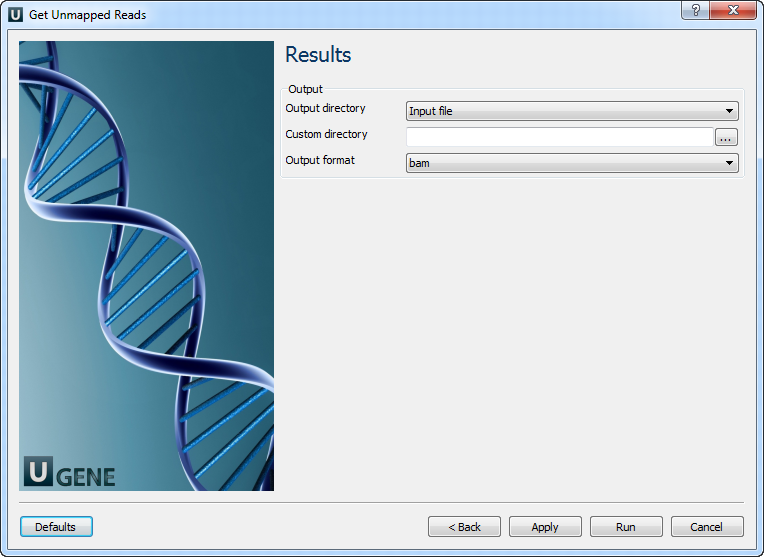
Results: On this page you need input output parameters.