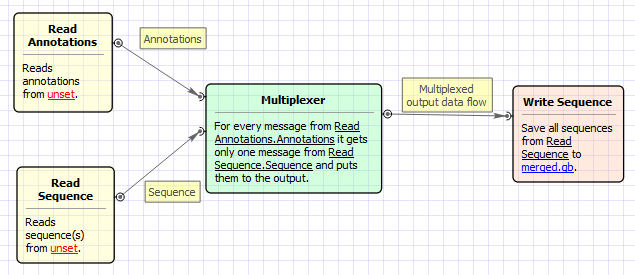
This sample workflow shows how to merge input sequences with sets of annotations.
For example, you may have sequences in FASTA format and annotations in GFF format, and you would like to merge them and save the result into GenBank files.
The steps of the workflow are these:
- The workflow reads sequences from the input sequence files, e.g. sequence1, sequence2, sequence3.
- The workflow reads annotations from the input files with annotations, e.g. ann_set1, ann_set2, ann_set3.
- The sequences and the annotations are multiplexed. The result is:
- sequence1 + ann_set1
- sequence2 + ann_set2
- sequence3 + ann_set3
- The result is written to the output files.
How to Use This Sample
If you haven't used the workflow samples in UGENE before, look at the "How to Use Sample Workflows" section of the documentation.
Workflow Sample Location
The workflow sample "Merge Sequences and Annotations" can be found in the "Scenarios" section of the Workflow Designer samples.
Workflow Image
The workflow looks as follows:
Workflow Wizard
The wizard has 3 pages.
Input sequence(s): On this page you must input sequence(s).
Input annotation(s): On this page you must input annotation(s).
Output data: On this page you can modify output parameters.