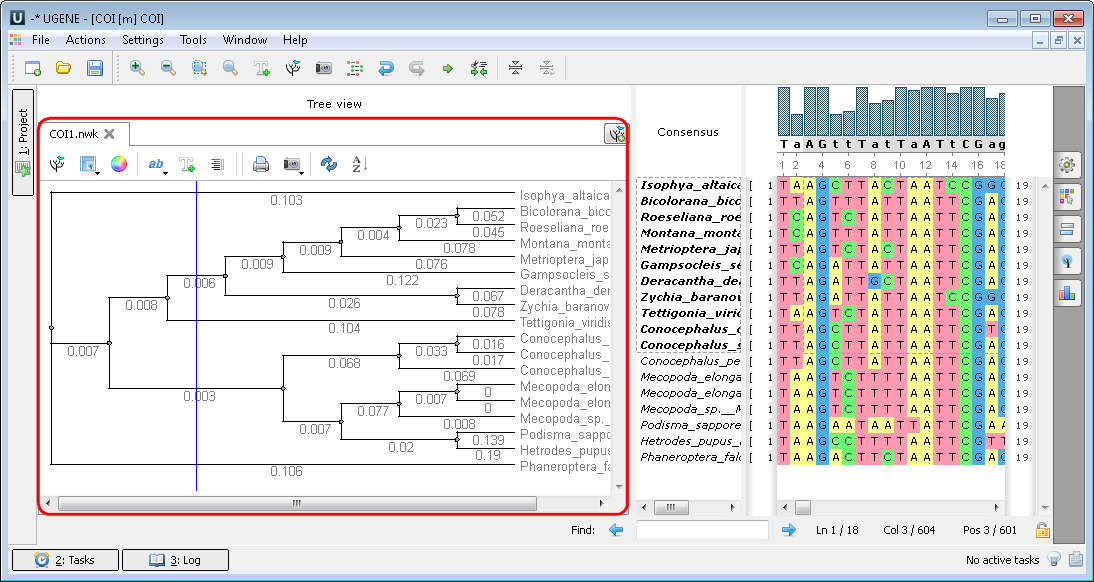
Multiple sequence alignment (MSA Editor). The Alignment Editor is a powerful tool for visualization and editing DNA, RNA or protein multiple sequence alignments. To activate the Alignment Editor open any alignment file. For example you can use the $ugene/data/samples/CLUSTALW/COI.aln file provided with UGENE. After opening the file in UGENE the Alignment Editor window appears:
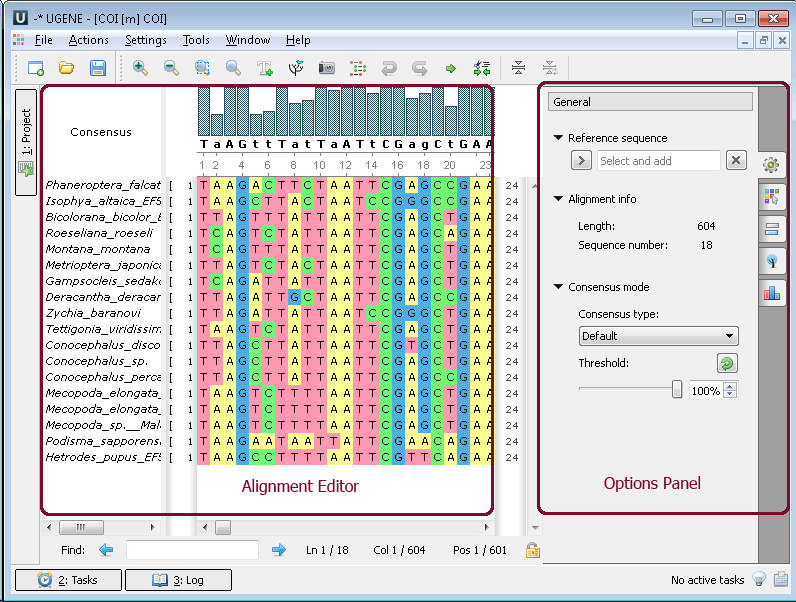
The editor supports different multiple sequence alignment (MSA) formats, such as ClustalW, MSF and Stockholm. The editor provides interactive visual representation which includes:
- Navigation through an alignment;
- Optional coloring schemes (for example Clustal, Jalview like, etc.);
- Flexible zooming for large alignments;
- Export publication-ready images of alignment;
- Several consensus calculation algorithms.
Using the Alignment Editor you can:
- Perform multiple sequence alignment using integrated MUSCLE and KAlign algorithms;
- Edit an alignment: delete/copy/paste symbols, sequences and subalignments;
- Build phylogenetic trees;
- Generate grid profiles;
- Build Hidden Markov Model profiles to use with HMM2/HMM3 tools.
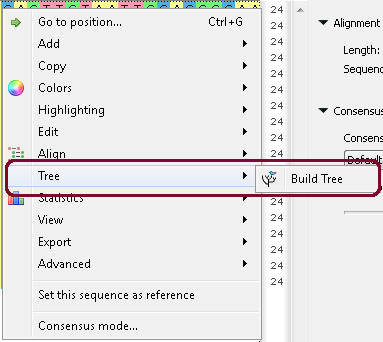
Example 2: Build a tree from your alignment. You can do this by three different ways:
a. From the toolbar. Click to the tree icon:

b. From the context menu:
c. From the Options Panel:
After calculation the tree appears in the MSA Editor in a separate window: