The Secondary Structure Prediction plugin provides a set of algorithms for the protein secondary structure (alpha-helix, beta-sheet) prediction from a raw sequence.
Currently, available algorithms are:
GORIV Jean Garnier, Jean-Francois Gibrat, and Barry Robson,”GOR Method for Predicting Protein Secondary Structure from Amino Acid Sequence”, in Methods in Enzymology, vol.266, pp. 540 - 553, (1996).
The improved version of the GOR method in J. Garnier, D. Osguthorpe, and B. Robson, J. Mol. Biol., vol. 120, p. 97 (1978).
PsiPred Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS. & Jones DT. (2005) Protein structure prediction servers at University College London. Nucl. Acids Res. 33(Web Server issue): W36-38.
Jones DT. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292: 195-202.
You can access these analysis capabilities for a protein sequence using the Analyze ‣ Predict secondary structure... context menu item. The dialog will appear:
It supports the following options:
Algorithm — you can choose the preferred algorithm. Currently, “GORIV” and “PsiPred” algorithms are available.
Range start / Range end — select the sequence range for prediction.
Results — the visual representation of the prediction results, for example:
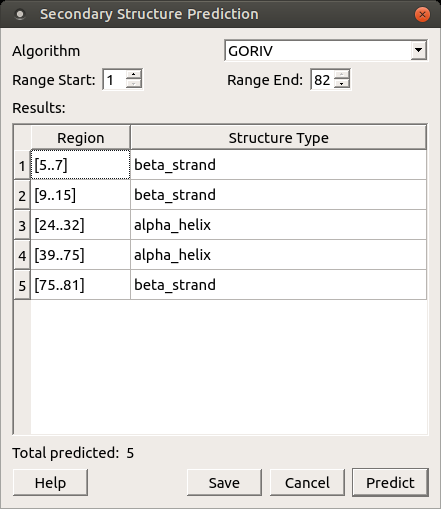
Save as annotation — select this button to save the results as annotations of the current protein sequence.