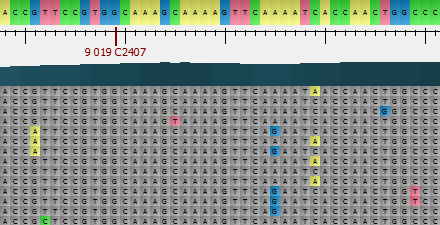
To apply an alignment highlighting mode, select it in the Highlighting context menu or on the Highlighting tab of the Options Panel. The following modes are available:
Agreements — .
Disagreements — highlights gaps and nucleotides that differ from the reference sequence.
- Strand direction — highlights reads located on the direct strand in blue and reads on the complement strand in green.
- Paired reads — highlights all paired reads in green. Note that the information about the pair is shown in the hint.