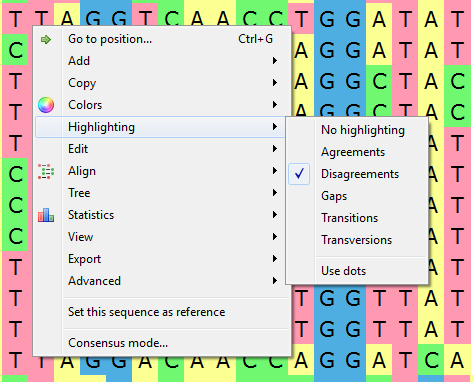
To apply an alignment highlighting mode, select it in the Highlighting context menu:
or on the Highlighting tab of the Options Panel:
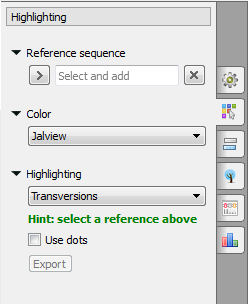
The following modes are available:
Agreements — .highlights symbols that coincide with the reference sequence.
Disagreements — highlights nucleotides that differ from the reference sequence.
- Gaps - highlights gaps.
- Transitions - highlights transitions.
- Transversions - highlights transversions.
To select a reference sequence use the Set this sequence as reference context menu or Reference sequence field in the Highlighting tab of the Options Panel.