From this chapter you can learn how to search for restriction sites on a DNA sequence.
The restriction sites found are stored as automatic annotations. This means that if the automatic annotations highlighting is enabled then the restiction sites are searched and highlighted for each nucleotide sequence opened. Refer Automatic Annotations Highlighting to learn more.
Open a DNA sequence in and click the following button on the Sequence View toolbar:
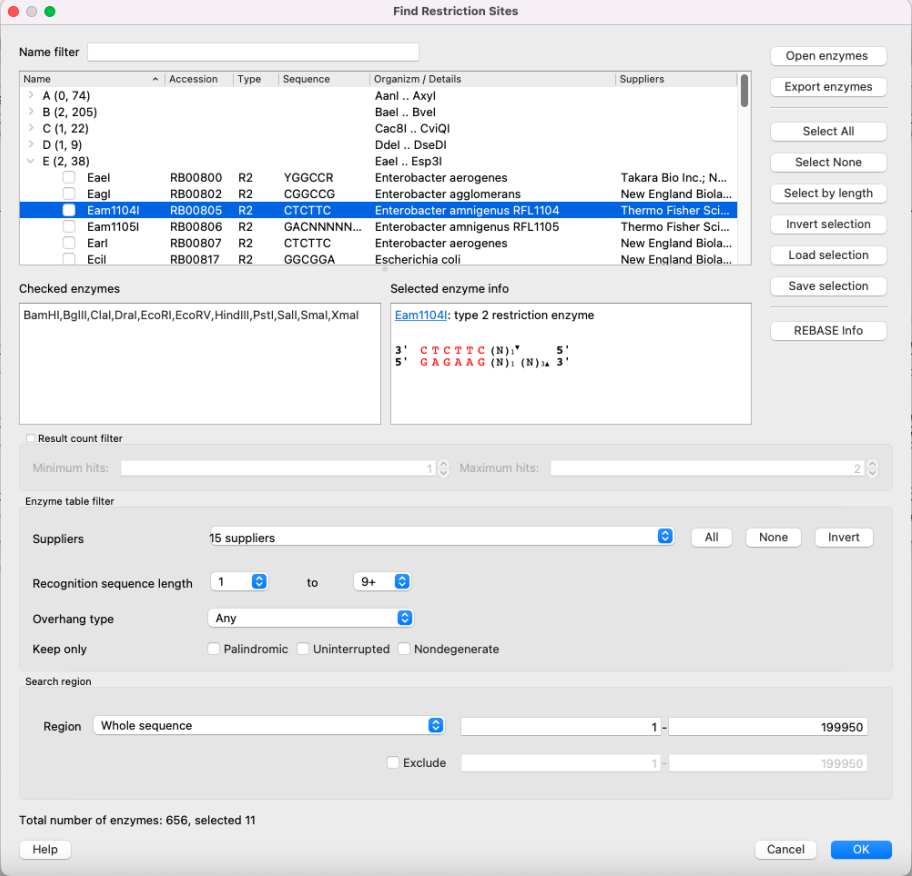
The Find restriction sites dialog appears:
The tree of restriction enzymes
Restriction enzymes are collected in a tree view. Enzymes are grouped by their names first letter here. Click on the > icon of a group (or just double click on the entire line) to show the whole group. Check the checkbox to include enzyme to the search list and uncheck to exclude. Use Name filter to find certain enzyme by its name.
Checked enzymes
Here you may see the all checked enzymes. This enzymes will be searched after clicking OK.
Selected enzyme info
Some information about the enzyme selected. It includes name, link to the REBASE database, the enzyme type detailed information, number of enzymes in the current sequence and sites and cuts location.
Control buttons (right side)
- Open enzymes - open file with enzymes in the bairoch format. It is required if you want to update enzymes database or open a database with some limited amount of enzymes. See Using Custom File with Enzymes for details.
- Export enzymes - export selected enzymes to the separate file (using the bairoch format).
- Select all - check all enzymes in the tree.
- Select none - uncheck all enzymes in the tree.
- Select by length - check enzymes only if its recognition site has length more than you set in the dialog window appeared.
- Invert selection - switch checked and unchecked enzymes.
- Load selection - load selected enzymes from the file. This file should contain comma-separated enzyme name list, for example: BamHI,BglII,ClaI,DraI,EcoRI,EcoRV,HindIII,PstI,SalI,SmaI,XmaI
- Save selection - save checked enzymes to the separate file using the format, described above.
- REBASE info - open the REBASE database page of the selected enzyme.
The filter of the results number
Show enzyme only if there are not less than "Minimum hints" and not more than "Maximum hints" values.
Enzyme table filter
Show/hide enzymes depending on certain parameters:
- Suppliers - check/uncheck suppliers, whose enzymes will be shown. By default, enzymes with undefined commercial suppliers are not shown.
- Recognition sequence length - show only enzymes, whose recognition sequence length is in bounds. NOTE: N bases are not included in recognition site. That means, that if enzyme has the following sequence:
C C N N N N N N N G G
its recognition site length will be four, not eleven. - Overhang type - show only enzymes with certain overhang type. The following options are presented:
- Any - show all enzymes.
- No overhang - site has no cuts at all. Example:
3' CTCGAG 5'
5' GAGCTC 3' - 5' - the cut of the forward strand is to the left of the cut of the reverse-complementary strand. Example:
3' C C▼T N A G G 5'
5' G G A N T▲C C 3' - 3' - the cut of the forward strand is to the right of the cut of the reverse-complementary strand. Example:
3' C G A T▼C G 5'
5' G C▲T A G C 3' - Blunt - the cut is located in the middle of the site.
- Sticky - the cut is locate anywhere but the middle of the site. It is 5' and 3' both.
- Nondegenerate sticky - the same as Sticky, but the overhang between cuts has only A, C, G or T (no extended) symbols.
- Blunt or sticky - Blunt + Sticky.
- Blunt or nondegenerate sticky - Blunt + Nondegenerate sticky.
- Keep only - show most interesting enzymes only:
- Palindromic - forward and reverse-complementary strands are equal.
- Uninterrupted - no internal N bases.
- Nondegenerate - No extended DNA alphabet symbols (only A, C, G, T and N).
Search region
The region to search enzymes in. You may also Exclude some region and not to search in it.
The information about enzymes was obtained from the REBASE database. For each enzyme in the list a brief description is available (the accession ID in the database, the recognition sequence, etc.). If you’re online you can get more detailed information about an enzyme selected by clicking the REBASE Info button.