The DNA Statistics plugin provides exportable statistic reports.
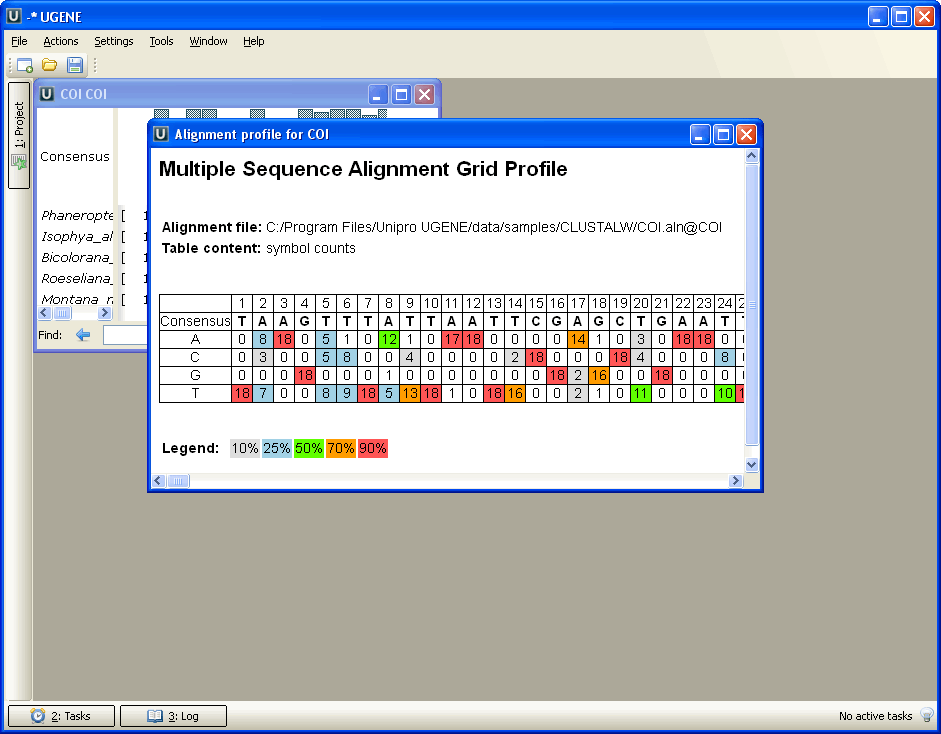
In the current UGENE version, the DNA Statistics plugin provides only Alignment Grid Profile report. The Alignment Grid Profile shows positional amino acid or nucleotide counts highlighted according to the frequency of symbols in a row.
The original idea of the MSA Grid Profile is described in the following paper:
“Alberto Roca, Albert Almada and Aaron C Abajian: ProfileGrids as a new visual representation of large multiple sequence alignments: a case study of the RecA protein family, BMC Bioinformatics 2008, 9:554”
Usage example:
Open a sequence alignment in the Alignment Editor and use the Statistics ‣ Generate grid profile context menu item.
The dialog will appear:
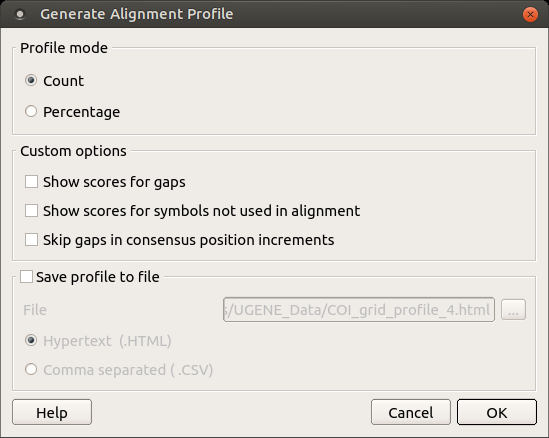
Here is a brief description of the options that can be set in the dialog:
Profile mode: Counts/Percents — select the Percents to have scores shown as percents in the report.
Show scores for gaps — check this item if you want gap characters (‘—’) statistics to be shown in the report.
Show scores for symbols not used in alignment — if a symbol is not used in the alignment at all it won’t be shown in the report. Check this item to make all symbols of alignment alphabet reported.
Skip gaps in consensus position increments — consensus ruler configuration. If checked the gaps in consensus will not lead to ruler increments.
Save profile to file — allows saving profile to a file in the HTML or CSV format. The CSV format is convenient for further processing in worksheets editors like Excel.
The resulting profile in the HTML mode: