The Annotations editor contains tools to manipulate annotations for a sequence. It provides a convenient way to organize, view and modify a single annotation as well as annotation groups.
An annotation for a sequence consists of:
- Name (or key) — indicates the biological nature of the annotated feature.
- Location — coordinates in the sequence.
- The list of qualifiers — qualifiers are the general mechanism for supplying information about annotation. Qualifiers are stored as pairs of (name, value) strings.
Below is the default layout of the Annotations editor with an extra column for the “note” qualifier added:
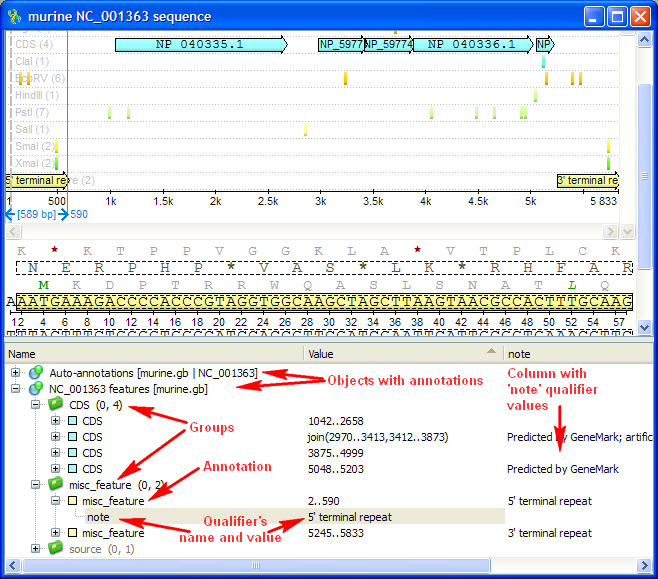
There are usually several objects with annotations in the Annotations editor. A special Auto-annotations object is always presented for each sequnce opened. It contains annotations automatically calculated for the sequence (see below for details).
An object contains groups of annotations used by UGENE for logical organization of the annotations. An annotation must always belongs to some group.
For documents created not by UGENE annotations are grouped by their names. For annotations created in UGENE it is possible to use arbitrary group names.
Groups can contain both annotations and other groups. The numbers in the brackets after a group name in the Annotations editor are the count of subgroups and annotations in the current group.
A single annotation is allowed to be presented in several groups simultaneously. An annotation is physically removed from the document when it does not belong to any group.