To create a DNA fragment from a sequence region activate the Sequence View window and select either the Actions ‣ Cloning ‣ Create Fragment item in the main menu or the Cloning ‣ Create Fragment item in the context menu.
The Create DNA Fragment dialog appears:
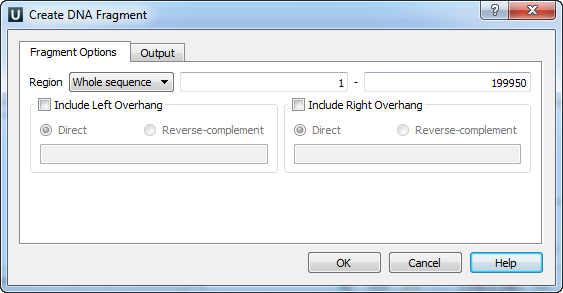
If a region has been selected you can choose to create the fragment from this region. Otherwise you can either choose to create the fragment from the whole sequence or choose the Custom item and input the custom region.
To add a 5’ overhang to the direct strand check the Include Left Overhang check box and input the required nucleotides. To add a 5’ overhang to the reverse strand in addition to the described steps select the Reverse-complement item in the same group box.
Similarly, to add a 3’ overhang check the Include Right Overhang check box, input the required overhang and select either the direct or the reverse-complement strand.
On the Output tab of the dialog you can optionally modify the annotations output settings.
Finally, press the OK button to create the fragment. The fragment will be saved as an annotation.