Download and install the UGENE FULL or NGS package to use this pipeline.
Use this workflow sample to process raw DNA-seq next-generation sequencing (NGS) data from the Illumina platform. The processing includes:
- Filtration:
- Filtering of the NGS short reads by the CASAVA 1.8 header;
- Trimming of the short reads by quality;
- Mapping:
- Mapping of the short reads to the specified reference sequence (the BWA-MEM tool is used in the sample);
- Post-filtration:
- Filtering of the aligned short reads by SAMtools to remove reads with low mapping quality, unpaired/unaligned reads;
- Removing of duplicated short reads.
The result filtered short reads assembly is provided in the SAM format. Intermediate data files are also available in the output.
How to Use This Sample
If you haven't used the workflow samples in UGENE before, look at the "How to Use Sample Workflows" section of the documentation.
Workflow Sample Location
The workflow sample "Raw DNA-Seq processing" can be found in the "NGS" section of the Workflow Designer samples.
Workflow Image
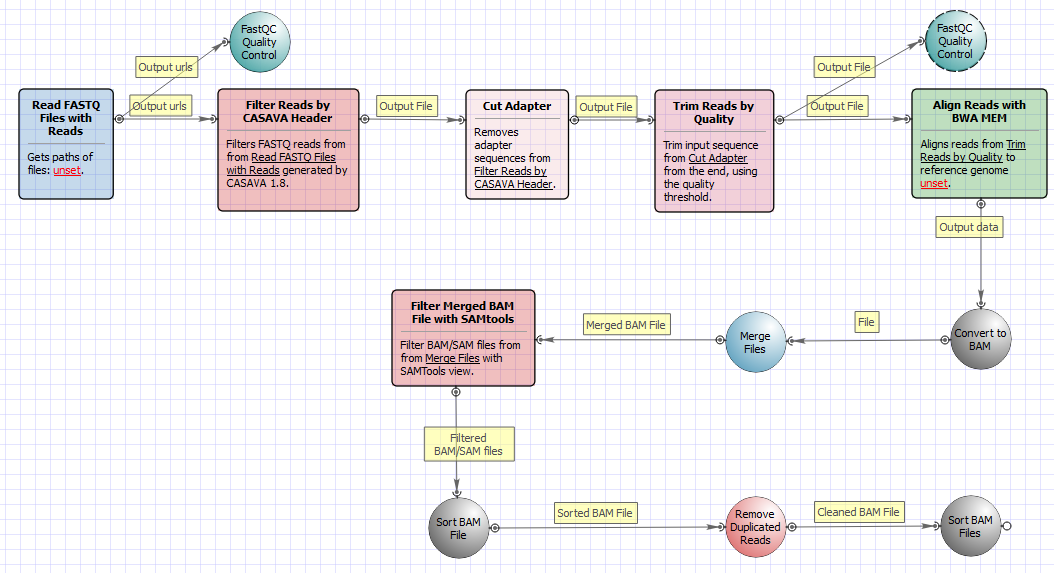
There are two versions of the workflow available. The workflow for single-end reads looks as follows:
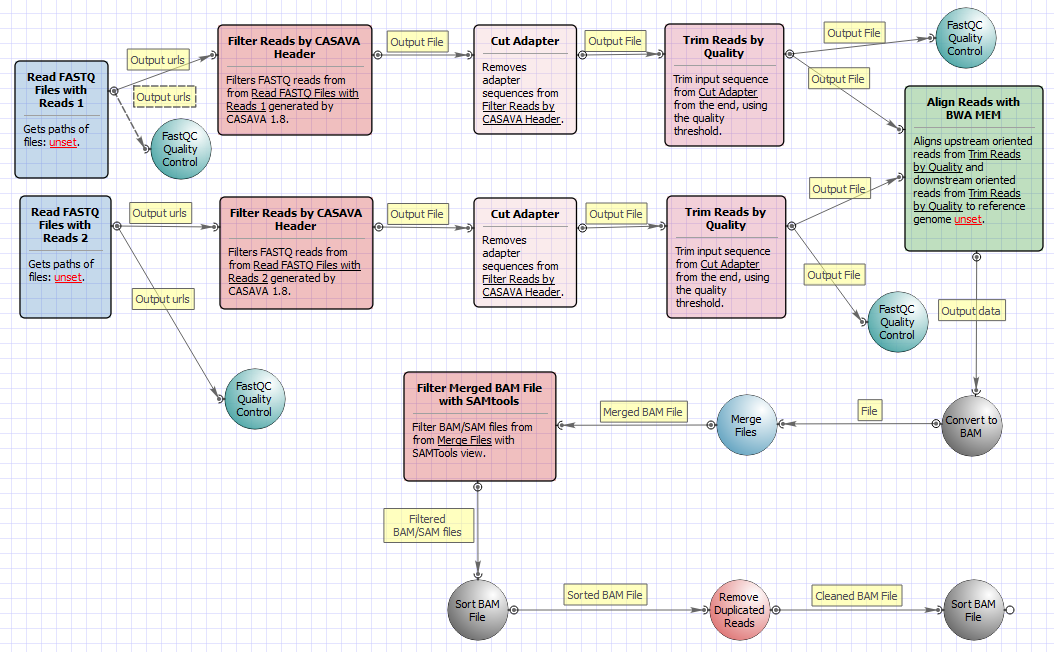
The workflow for paired-end short appearance is the following:
Workflow Wizard
The wizard for single-end reads has 5 page.
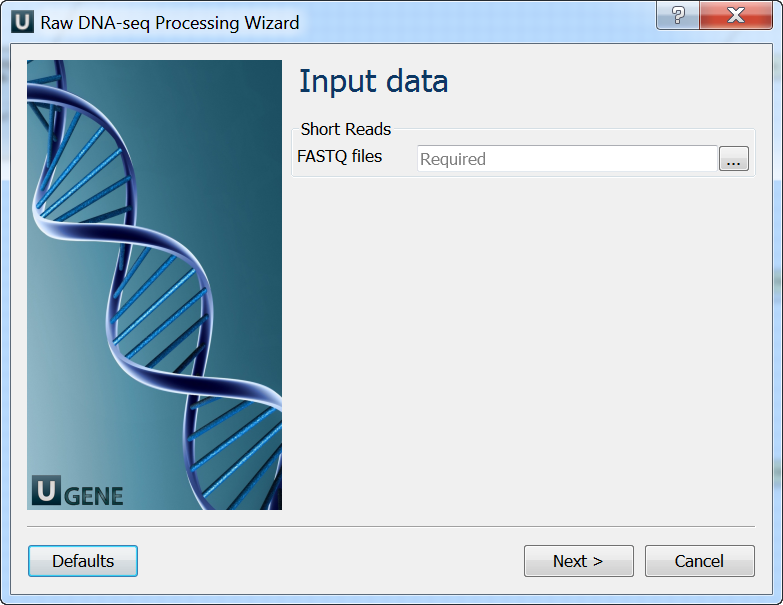
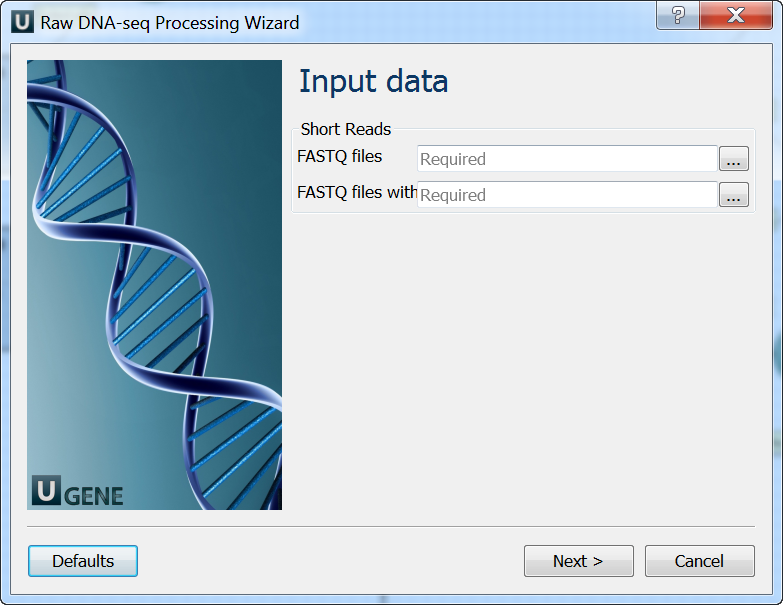
Input data: On this page you must input FASTQ file(s).
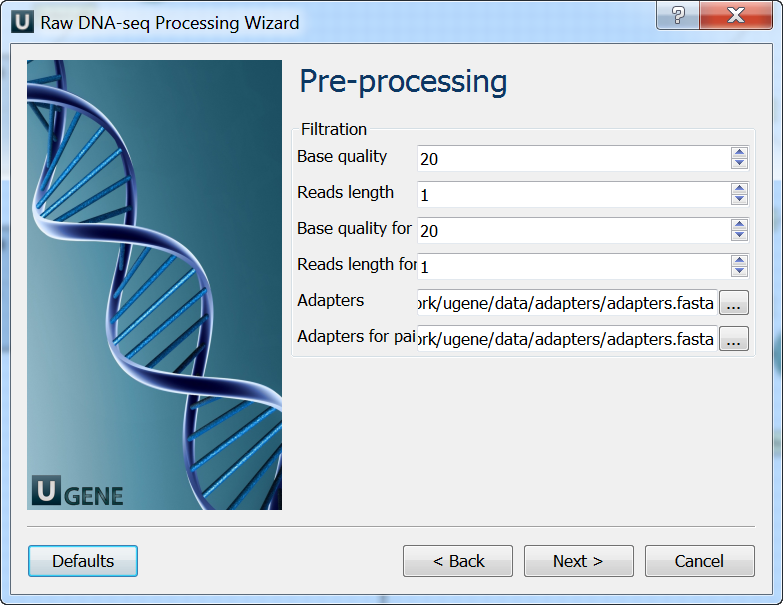
Pre-processing: On this page you can modify filtration parameters.
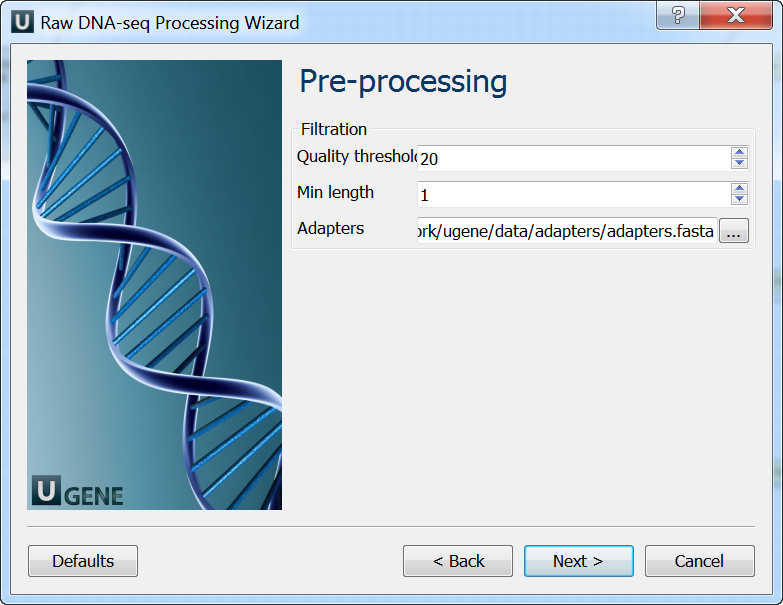
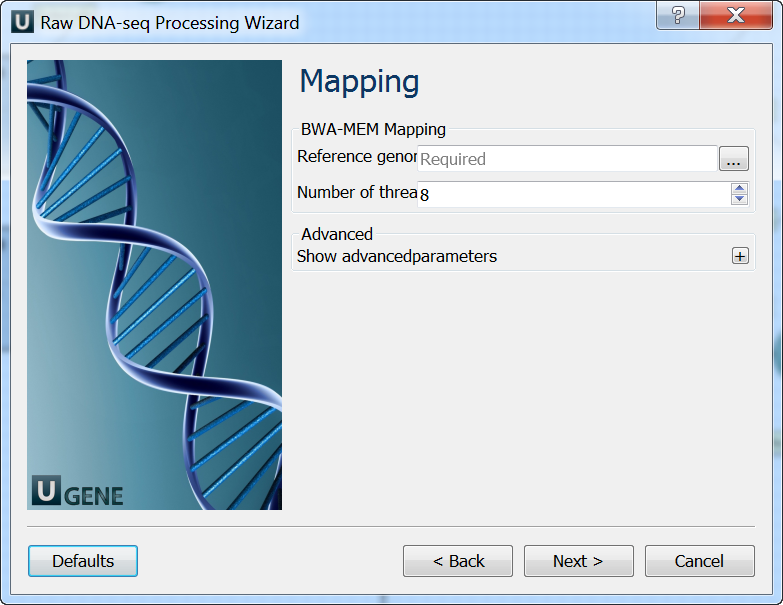
Mapping: On this page you must input reference and optionally modify advanced parameters.
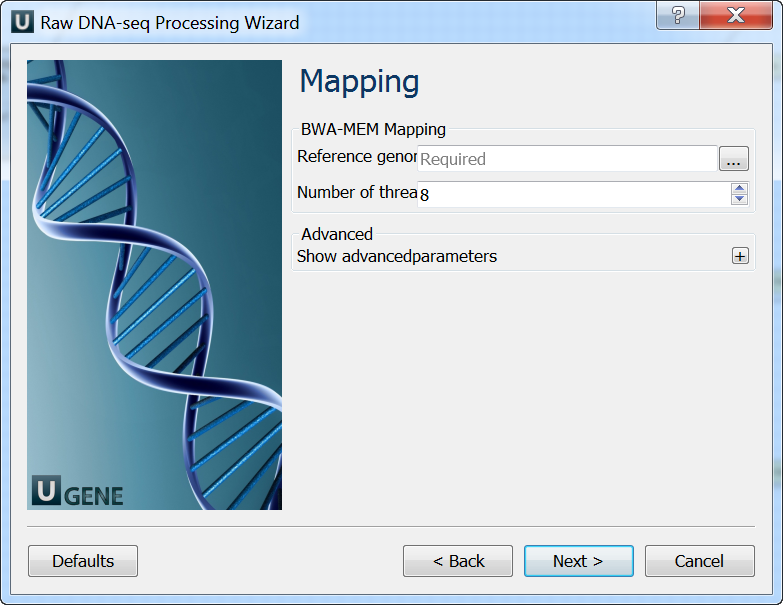
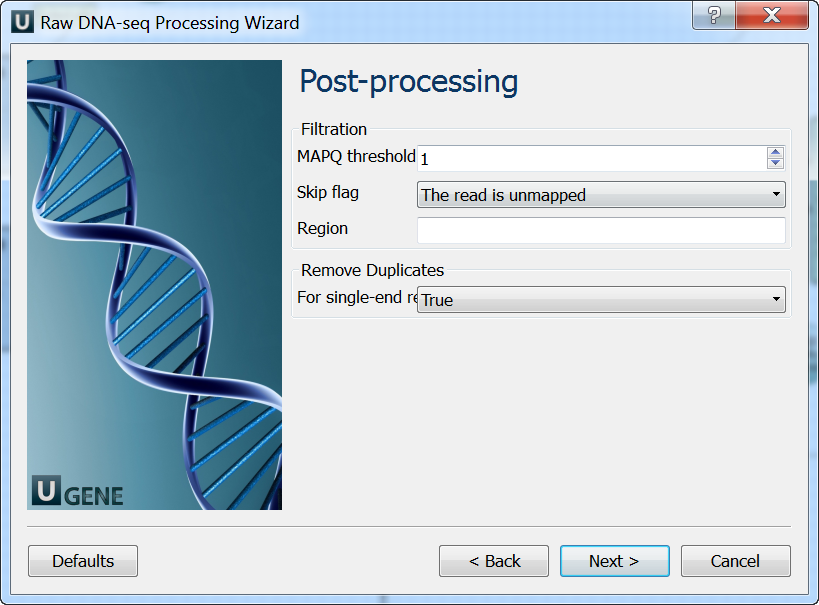
Post-processing: On this page you can modify post-processing parameters.
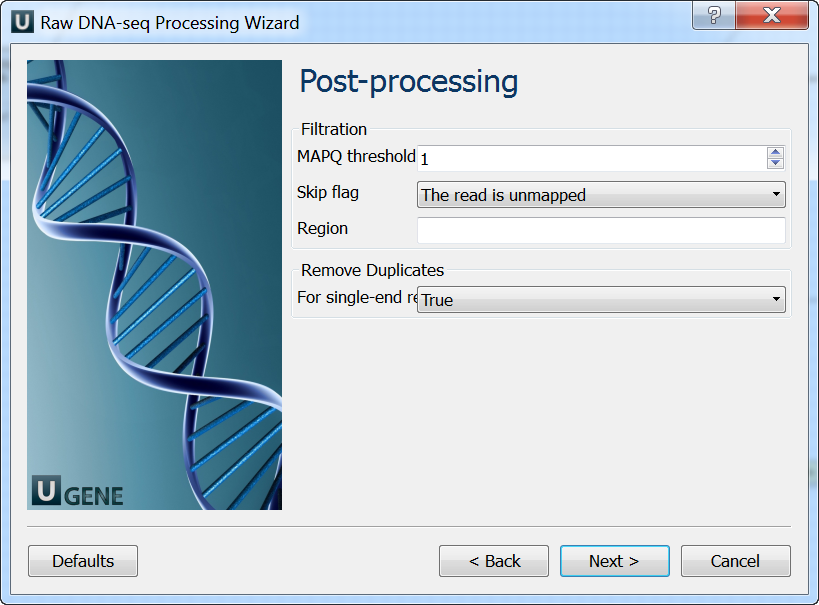
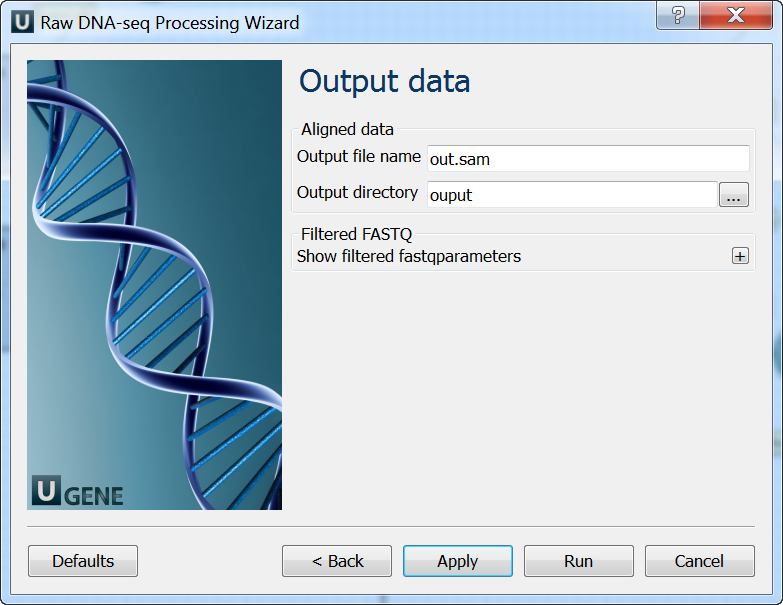
Output data: On this page you must input output parameters.
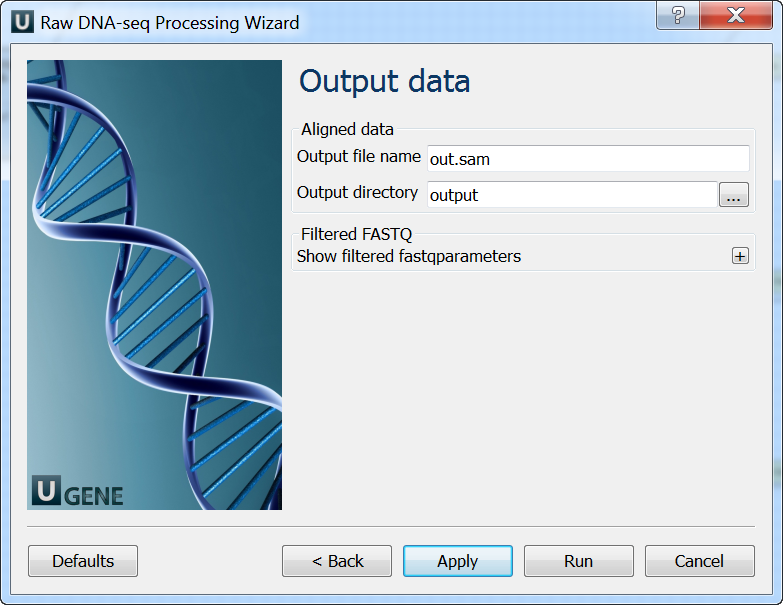
The wizard for paired-end reads has 5 page.
Input data: On this page you must input FASTQ file(s).
Pre-processing: On this page you can modify filtration parameters.
Mapping: On this page you must input reference and optionally modify advanced parameters.
Post-processing: On this page you can modify post-processing parameters.
Output data: On this page you must input output parameters.