SnpEff is a variant annotation and effect prediction tool. It annotates and predicts the effects of genetic variants (such as amino acid changes).
A typical SnpEff use case would be:
-Input: The inputs are predicted variants (SNPs, insertions, deletions and MNPs). The input file is usually obtained as a result of a sequencing experiment, and it is usually in variant call format (VCF).
-Output: SnpEff analyzes the input variants. It annotates the variants and calculates the effects they produce on known genes (e.g. amino acid changes).
How to Use This Sample
If you haven't used the workflow samples in UGENE before, look at the "How to Use Sample Workflows" section of the documentation.
Workflow Sample Location
The workflow sample "Variation Annotation with SnpEff" can be found in the "NGS" section of the Workflow Designer samples.
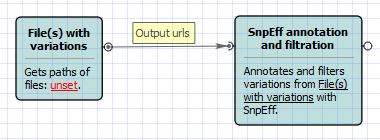
Workflow Image
The opened workflow looks as follows:
Workflow Wizard
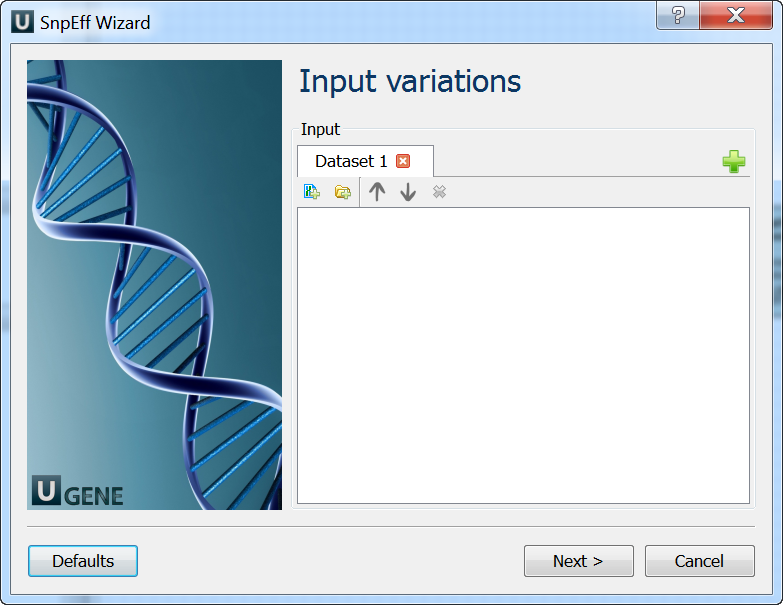
The wizard has 3 pages.
Input Variations: On this page you must input variations file(s).
- SnpEff Parameters: On this page you can modify SnpEff parameters.
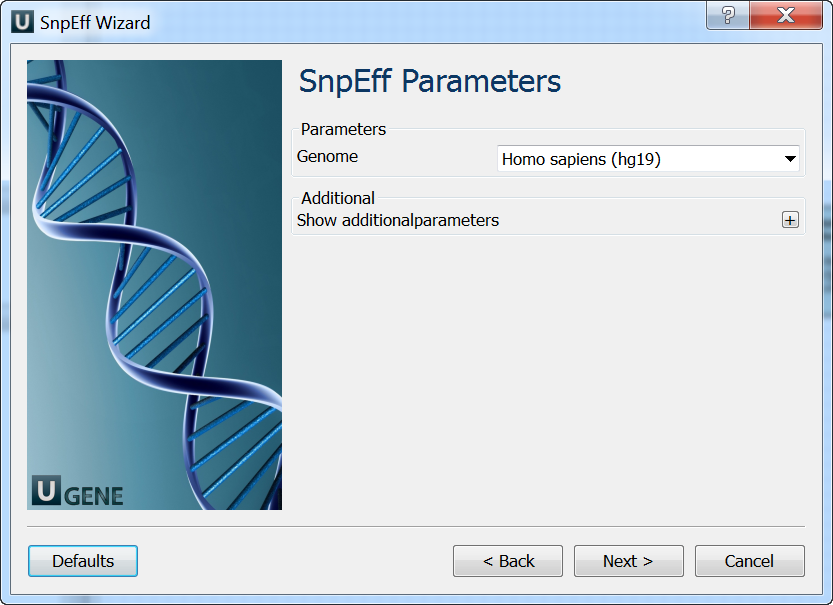
- Output: On this page you need input output parameters.