...
Tool home page: mfold.org.
| Anchor | ||||
|---|---|---|---|---|
|
UGENE uses Ghostscript to convert PS files to PNG and PDF.
ToC
| Table of Contents | ||
|---|---|---|
|
...
| Parameter | Default | Description | ||||
|---|---|---|---|---|---|---|
| Save output to | /path/to/sequence/ | The folder where the "mfold" subdirectory will be created with the output data in it. By default, this folder is the same as the folder where the input sequence is stored. For example, let's say our OS is Windows and the analyzed sequence has the path "C:\path\to\sequence\my_sequence.fa". After running the task, the folder structure will look something like this
| ||||
| 96 | Setting up the Ghostscript converter from ps PS files to PNG. Quality of saved images (PNG files). The higher this parameter, the higher the quality, size and resolution of the resulting images.
|
...
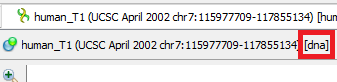
- Molecule Type (DNA/RNA). mfold gets sequence information from command line arguments. Information about what type will be passed to the tool can be seen next to the sequence name
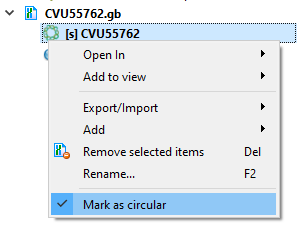
- Molecule Topology (linear/circular). mfold gets sequence information from command line arguments. If the sequence is marked as circular, then mfold will work with it as a circular one. Otherwise, as with linear one.
Whether the sequence is linear/circular is described here.
Part of a circular sequence will also be considered a circular sequence. If you want to change this behavior, uncheck "Mark sequence as circular" and then call the mfold dialog - DPI of images in UGENE. Ghostscript has a default DPI of 72. Therefore, this value is used for images for the internal UGENE report and this value cannot be changed.
...
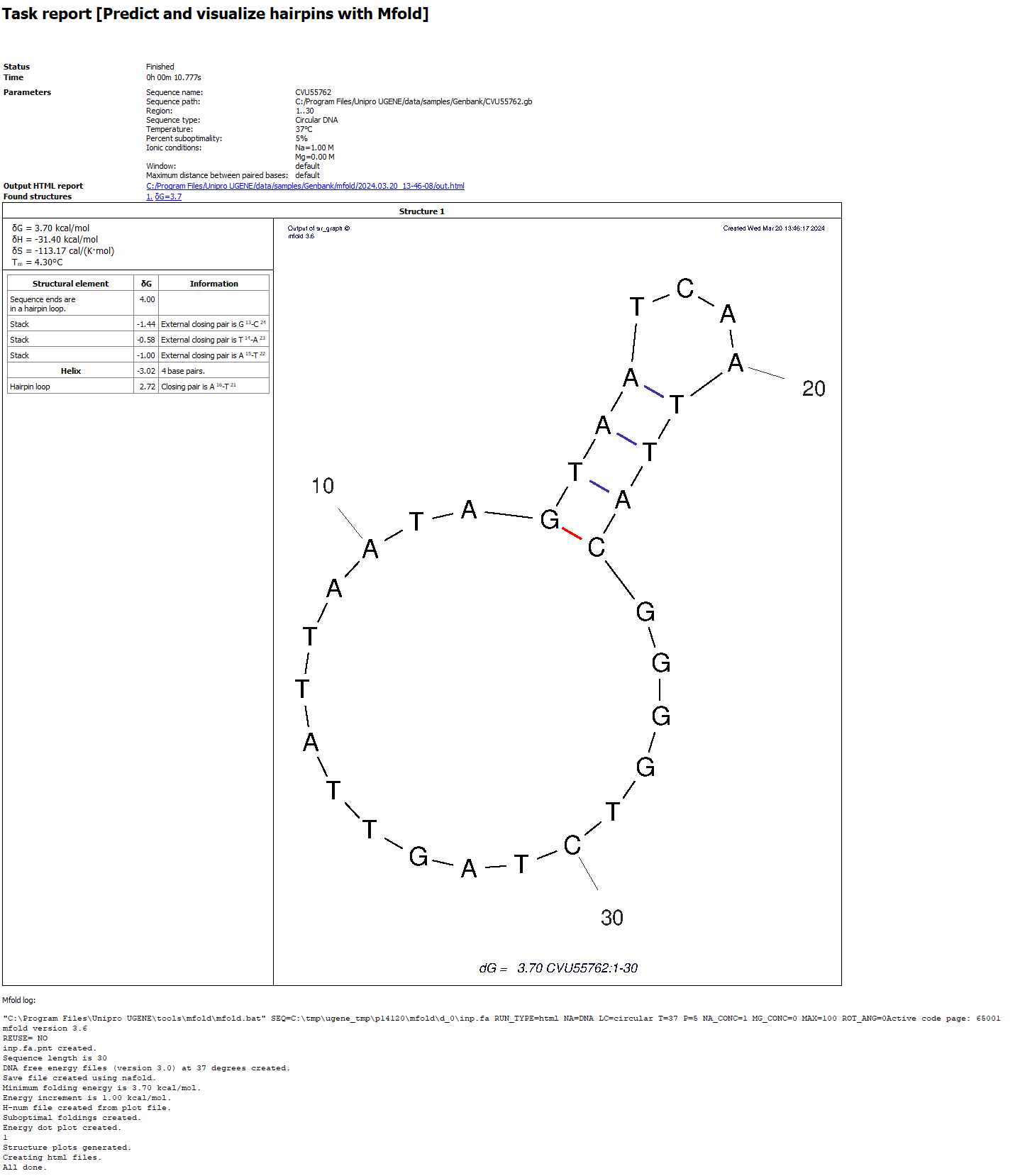
Example of a report on Windows (appearance may vary slightly between systems):
- Status, Time, … At the beginning there is information about the run: information about the task, settings for launch.
- Output HTML report. Clickable link to the output HTML report. More about this report in the next section.
- Found structures. List of found structures. The structure is characterized by free energy. When clicked, UGENE will scroll the window to the desired structure.
- Structure table. The table contains
- Thermodynamic details: free energy, enthalpy, entropy and an estimated Tm.
- Loop Free-Energy Decomposition: the entire decomposition of the particular folding into loops and stacks, together with their free energies and closing base pairs. Consecutive runs of base pairs are summarized as helices.
- Image. Displays of nucleic acid secondary structure. At the bottom of the image there is information identifying it: free energy, name of the input sequence and region of analysis.
- mfold log. Information about launching the external mfold tool: launch command and log.
...
| Info |
|---|
Sometimes the report may not contain the structure table. This happens because there are a lot of results/the resulting decomposition tables are too large. In this case, the result must be viewed in a browser (click on the "Output HTML report" link). |
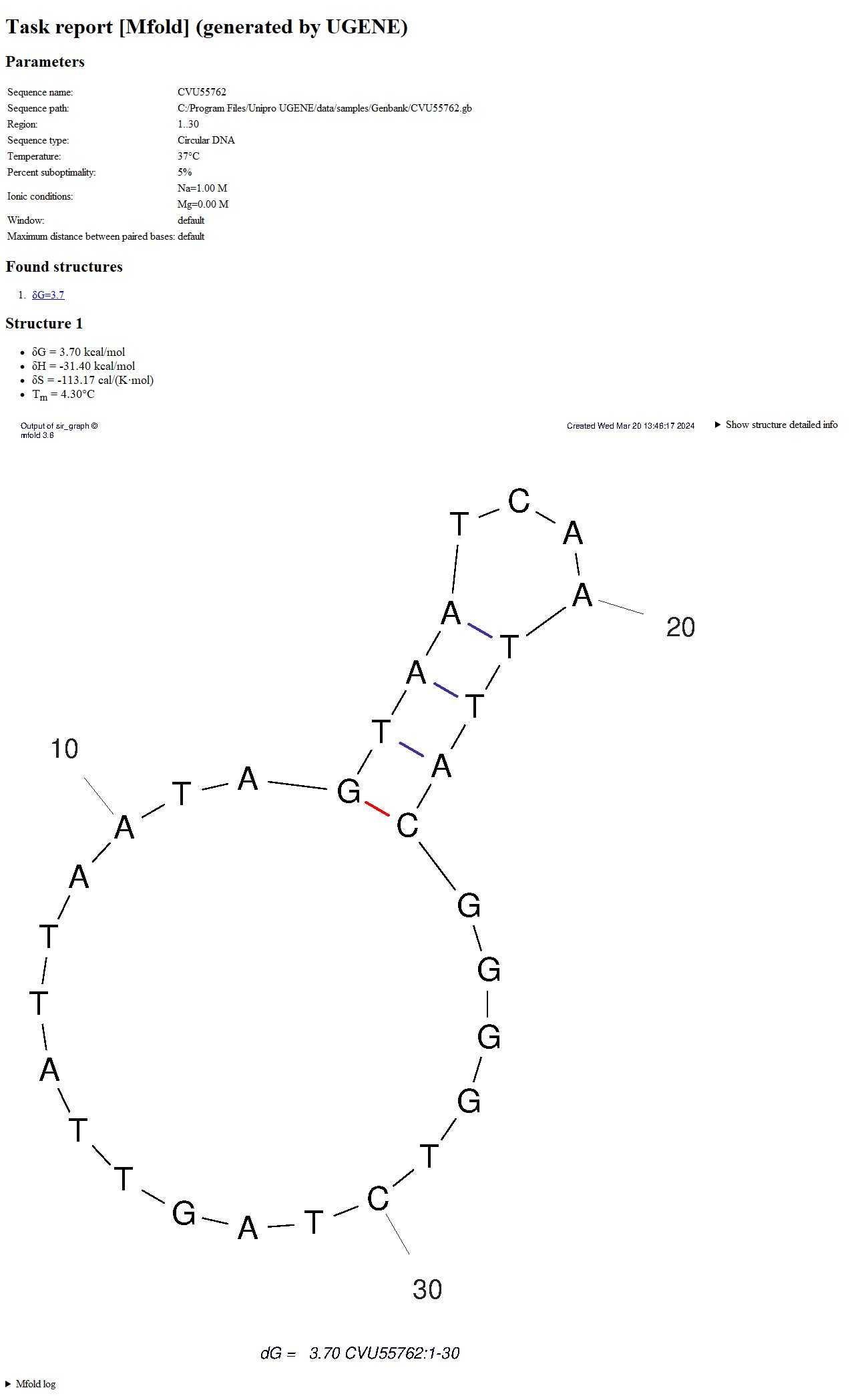
HTML report
A single HTML report and PNG+PDF for each structure found are saved in the output folder. The HTML report displays these images, PDFs and detailed information about the launch in an easy-to-view form.
Tool comparison
хранение результатов
подключение к интернету
какие настройки есть
исправлены баги
размер входных последовательностей
LinksThis version of the report also contains:
- Launch information.
- List of found structures with links to them.
- Information about each structure. The Loop Free-Energy Decomposition is located under the "Show structure detailed info" arrow.
The quality of the images in the report depends on the DPI setting in the dialog. When you click on the picture, a PDF file with a structure image opens in a new tab, identical to the picture image, but allows you to zoom without losing quality. - mfold log.
This report is portable, i.e., if the directory is completely copied (HTML+all PNGs+all PDFs), the copied data can be re-opened in the browser without loss of functionality. Folders can be deleted/renamed; it will not affect the operation of UGENE.
Temporary output
| Info |
|---|
For advanced users. |
mfold produces many files. Not all of them are displayed in the reports. However, while the current UGENE session is open, you can see what other files mfold produces.
In the Log corresponding to the [Algorithms][DETAILS] subcategories, you can see the mfold launch command. It depends on where the UGENE temporary directory is set. For example, it might look like this
| Code Block |
|---|
"C:\Program Files\Unipro UGENE\tools\mfold\mfold.bat" SEQ=C:\tmp\ugene_tmp\p14120\mfold\d_0\inp.fa RUN_TYPE=html NA=DNA LC=circular T=37 P=5 NA_CONC=1 MG_CONC=0 MAX=100 ROT_ANG=0 |
In this case, all temporary output files will be located in C:\tmp\ugene_tmp\p14120\mfold\d_0. For information about the purpose and format of this data, please read the article.
| Warning |
|---|
| Do not modify/move/delete these files. They are read-only. |
Tool comparison
As mentioned earlier, the mfold tool can also be launched from the native site or from the IDT platform.
| UGENE | unafold.org | OligoAnalyzer | |
|---|---|---|---|
| Internet connection | not required | must be (http connection) | must be |
| Privacy | share only what you want | results are visible to everyone | results are visible only to you |
| Storing results | stored as long as needed | are removed over time | stored while the session is open (30 minutes) |
| File output completeness | no energy dot plot | there are all files including energy dot plot | there is energy dot plot but not some other side files |
| Image quality | vector format is represented by PDF files quality of raster images (PNG) is configurable | vector format is represented by PDF and PS files quality of raster images (PNG, JPEG) is configurable | no vector format raster images aren't configurable |
| Completeness of algorithm settings | some settings for RNA aren't taken into account no constraints | all constraints can be specified | truncated part no constraints, have their own settings |
| Completeness of display settings | impossible to specify region's offset relative to original sequence | offset setting | no offset |
| Max sequence length | 3000 | 2400 | 255 |
https://en.wikipedia.org/wiki/List_of_RNA_structure_prediction_software
References
- Article https://www.ncbi.nlm.nih.gov/pmc/articles/PMC169194/
- mfold home http://www.unafold.org/
- mfold DNA http://www.unafold.org/mfold/applications/dna-folding-form.php
- mfold RNA http://www.unafold.org/mfold/applications/rna-folding-form.php
- mfold sources http://www.unafold.org/mfold/software/download-mfold.php
- mfold settings documentation http://www.unafold.org/mfold/documentation/mfold-documentation.php
- Viewing launch results http://www.unafold.org/cgi-bin/view-folds.cgi
- Forum with mfold authors http://www.unafold.org/forum/
- mfold references http://www.unafold.org/mfold/documentation/mfold-references.php
- implementation of mfold by IDT company https://www.idtdna.com/calc/analyzer