...
If you set "5' backbone length", then the number of bases you set from 5' end of the backbone sequence will be checked for unwanted connections. The same about "3' backbone length", but from the 3' end. If the checked end has hairpins, self- and homo-dimers you will see the following dialog:
| Children Display | ||
|---|---|---|
|
Type two primers for running In Silico PCR. If the primers pair is invalid for running the PCR process then the warning is shown. Also, primers for the running In silico PCR can be chosen from a primer library. Click the following button to choose a primer from the primers library:
| HTML |
|---|
<center>
<br>
<img src="/wiki/download/attachments/13435073/Primers Library.png"/>
</br>
</center> |
The following dialog will appear:
| HTML |
|---|
<center>
<br>
<img src="/wiki/download/attachments/16125631/In Silico PCR_2.png"/>
</br>
</center> |
The table consists of the following columns: name, GC-content (%), Tm, Length (bp) and sequence. Select primer in the table and click the Choose button.
Click the Reverse-complement button for making a primer sequence reverse-complement:
| HTML |
|---|
<center>
<br>
<img src="/wiki/download/attachments/13435069/In Silico PCR_1.png"/>
</br>
</center> |
Click Show primers details for seeing statistic details about primers.
When you run the process, the predicted PCR products appear in the products table.
Products table
There are three columns in the table:
- region of product in the sequence
- product length
- preferred annealing temperature
Click the product for navigating to its region in the sequence.
Click the Extract product(s) button for exporting a product(s) in a file or use double click for that.
...
Other sequences in PCR reactions
Sequences that are supposed to be in the reaction mixture. Its important primers do not have unwanted connections to these sequences, so it will be checked that there are no hairpins, self- or hetero-dimers between the resulting primer and any sequence from the "Other sequences in PCR reaction" file.
Result
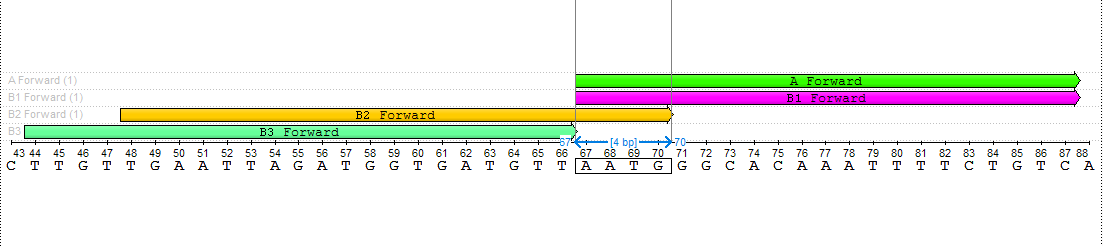
The search result is from 1 to 4 pairs of primers. The results should be as close as possible to the amplified fragment. That means, that the searching process starts from the edge of the amplified fragment to the 5' direction on the direct strand in case of the forward primer and to the 3' direction of the reverse-complementary strand in case of the reverse primer. The results are defined with special names:
- A - the pair of primers closest to the amplified fragment, corresponding to the entire set of parameters.
- B1 - the pair of primers closest to the amplified fragment, corresponding to the entire set of parameters if B2 and/or B3 exist.
- B2 - the pair of primers corresponding to the entire set of parameters, which has a 4-nucleotide intersection with B1.
- B3 - the pair of primers corresponding to the entire set of parameters, which starts immediately when B1 ends.
Also, you may see results in the result table. A single click on the result in the result table selects the corresponding annotation, double click - export the resulting primer to the separate file (with the backbone sequence, which will be added during the exporting process).
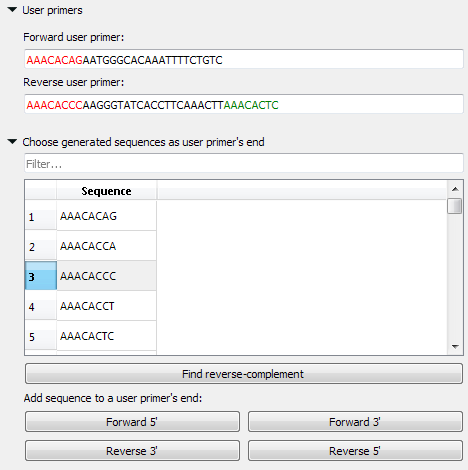
User primer
You may check the validity of the pair of primers by typing this pair into the corresponding fields. Also, you may add your primers 5' or/and 3' 8 bases endings, which do not form simple unwanted connections.