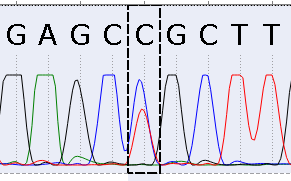
Some mutations could have a chromatogram signal lesser than the reference one. This situation very often appears if the mix of cells (healthy and oncological) has been being analyzed. That means that the mutation signal could be different (due to a different concentration of oncological cells in a sample).
Look at the picture below. The signal of the "C" nucleotide (blue) is the highest. But there is a signal of the "T" nucleotide (red), which is high enough too. The second strongest signal is the thing we call Alternative mutation.
It's very important to make Alternative mutations clearly visible. To do this, you can use the Alternative mutations function.
Show alternative mutations -
You can change font by Appearance–>Change characters font context menu item or by the main Actions–>Appearance menu item.