SPAdes – St. Petersburg genome assembler. Click this link to open SPAdes homepage. SPAdes is embedded as an external tool into UGENE.
Open Tools ‣ NGS data analysis.
Select the Genome de novo assembly item to use the SPAdes.
The Assemble Genomes dialog will appear.
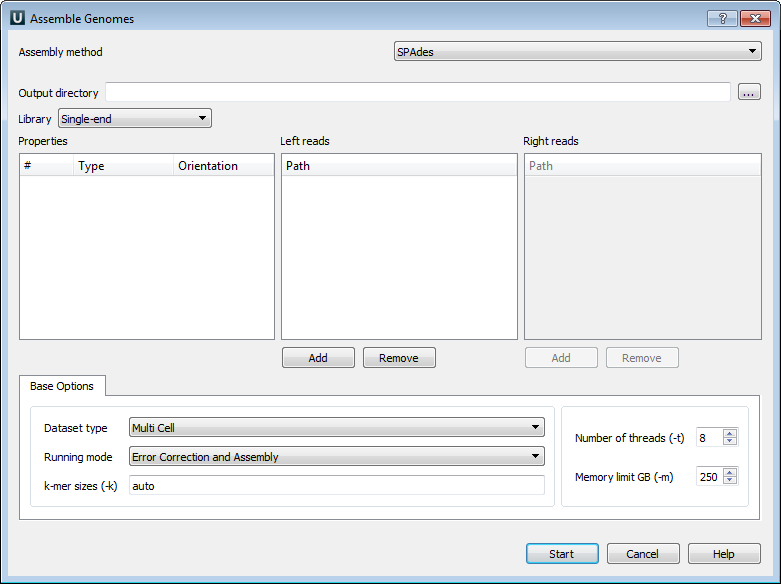
The following parameters are available:
Output directory - SPAdes stores all output files in output directory, which is set by the user.
Library - to run SPAdes choose one of the following libraries:
- Single-end
- Paired-end
- Paired-end (Interplaced)
- Paired-end (Unpaired files)
- Sanger
- PacBio
Left reads - file(s) with left reads.
Right reads - file(s) with right reads.
For each dataset in the paired-end libraries you can change type and orientation.
Datasest type - dataset type.
Running mode - running mode.
k-mer sizes (-k) - k-mer sizes.
Number of threads (-t) - number of threads.
Memory limit GB (-m) - memory limit.