Plasmid Auto Annotation feature allows to automatically annotate possible functional elements of the given sequence such as promoters, terminators, origin of replication, known genes, common primers and other features. Conceptually this functionality is similar to the one offered by PlasMapper software. The database for plasmid auto-annotation is based on the following resource.
To activate Plasmid Auto Annotation upon your sequence use the menu item Analyze ‣ Annotate plasmid. In the appeared dialog one can selected the features to search in sequence.
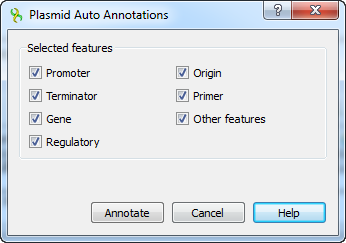
The detected plasmid features are stored as automatic annotations and can be controlled through corresponding menu. Refer Automatic Annotations Highlighting to learn more.
Plasmid Auto Annotations consists 471 carefully selected and annotated plasmid features broken down into 12 feature categories.
FeatureDB consists of a single FASTA file with These are shown in Table 1. FeatureDB was created by combining the original FASTA formatted PlasMapper 2.0 feature database (which had 336
features) with AddGene’s sequence feature database (https:
//www.addgene.org/) followed by extensive manual editing,
duplicate removal, validating and cleaning.